Abstract
Background
SNP genotyping has become increasingly more common place to understand the genetic basis of complex diseases like cancer. SNP-genotyping through MassARRAY™ is a cost-effective method to quantitatively analyse the variation of gene expression in multiple samples, making it a potential tool to identify the underlying causes of colorectal carcinogenesis.
Methods
In the present study, SNP genotyping was carried out using Agena MassARRAY™, which is a cost-effective, robust, and sensitive method to analyse multiple SNPs simultaneously. We analysed 7 genes in 492 samples (100 cases and 392 controls) associated with CRC within the population of Jammu and Kashmir. These SNPs were selected based on their association with multiple cancers in literature.
Results
This is the first study to explore these SNPs with colorectal cancer within the J&K population.7 SNPs with a call rate of 90% were selected for the study. Out of these, five SNPs rs2234593, rs1799966, rs2229080, rs8034191, rs1042522 were found to be significantly associated with the current study under the allelic model with an Odds Ratio OR = 2.981(1.731–5.136 at 95% CI); p value = 4.81E-05 for rs2234593,OR = 1.685(1.073–2.647 at 95% CI);; p value = 0.02292 for rs1799966, OR = 1.5 (1.1–2.3 at 95% CI), p value = 0.02 for rs2229080, OR = 1.699(1.035–2.791 at 95% CI); p value = 0.03521 for rs8034191, OR = 20.07 (11.26–35.75); p value = 1.84E-34 for rs1042522 respectively.
Conclusion
This is the first study to find the relation of Genetic variants with the colorectal cancer within the studied population using high throughput MassARRAY™ technology. It is further anticipated that the variants should be evaluated in other population groups that may aid in understanding the genetic complexity and bridge the missing heritability.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is the third most common type of cancer worldwide, resulting in 1–2 million new cases each year (1). In 2020, it was estimated that nearly 10% of all cancer incidences were reported to be of CRC [1]. The incidence of CRC has been associated with obesity, red meat consumption, and physical inactivity [2, 3]. In addition, the genetic factors and epigenetic changes also play a key role in the initiation and progression of CRC [4, 5]. Delay in the diagnosis of CRC is a major hurdle in the management of CRC, which is evident by the rise in new cases each year. Therefore, it is critical to identify markers that may help in the early prognosis and development of therapeutic interventions accordingly.
In India, CRC accounted for about 1,479,783 deaths in the year 2020 [1]. One recent study has put Colorectal cancer as the forth most common cancer in the Jammu and Kashmir (J&K) region [6], there is a spike in the incidence of gastric, oesophagus and CRC in recent years. The rise in the incidence of these disorders may be as a result of lifestyle and food consumption [7]. However, the genetic aspect of gastro-intestinal cancer cannot be overruled (11). According to the J&K based hospital report, CRC accounted for the forth most common type of cancer [6]. In this regard, identification of the CRC related genetic variants in the region of J&K is necessary for the proper prognosis and CRC management, which can be achieved through single-nucleotide polymorphisms (SNP) genotyping. Several studies have looked at mutations in critical genes involved in cell cycle, cell growth, DNA damage repair, and a variety of other processes in the J&K population with regard to various malignancies [8,9,10].
SNP genotyping is a powerful tool that has identified the genetic basis of complex disease, including CRC [11]. The studied SNPs hence provides key insights on the molecular pathogenesis of cancer that can be further translated to the identification of cancer and therapeutic biomarkers [12,13,14]. Identifying the role of these genetic variants may provide valuable insights on prognosis and optimize therapies for the treatment of CRC. The Agena Bioscience MassARRAY™ System provides genotype data for several user-defined SNPs in a large number of DNA samples in a high-throughput and cost-effective manner [11]. In this study, we have carried out SNP genotyping using Agena MassARRAY™ to identify multiple SNPs and samples simultaneously.
The previous study in Chinese population has showed that rs2229080 of DCC (Deleted in Colorectal Carcinoma Netrin1 Receptor)was associated with low breast cancer risk [15]. In contrast, DCC rs2229080 displays no significant association with esophageal cancer risk in the region of J&K [16]. In the present study, we analysed 7 genes and found out that rs2234593, rs1799966, rs2229080, rs8034191, rs1042522 were found to be significantly associated with colorectal cancer in the current studied population of J&K region. Taken together, our study investigated the role of cancer-related genetic variants in CRC in the population of J&K. The study of these genetic variants may provide valuable insights on the proper prognosis and CRC management. However, more large-scale sample size studies are required that will further support the present study.
Material and methods
Sample collection
This study was approved by the Institutional Ethics Review Board (IERB) of Shri Mata Vaishno Devi University (SMVDU). All details were recorded in a pre-designed proforma and the written informed consent was obtained from each participant before conducting the study.
A total of 492 participants including 100 colorectal cancer patients and 392 healthy controls (age and sex-matched) were recruited from the Jammu and Kashmir region of India. All the participants recruited for the study were obtained from hospitals and various clinics of J&K. 2 ml of venous blood was collected in ethylenediaminetetraacetic acid (EDTA) vacutainer tubes from all the participants. The clinical parameters of both cases and controls are provided in Supplementary Table 1.
DNA extraction
The genomic DNA was isolated from the blood samples, using the manufacturer’s protocol of Qiagen™ DNA isolation Kit (Catalogue No. #51206, Hilden, Germany). Genomic DNA was quantified using Eppendorf’s Bio Spectrometer™ (Hamburg, Germany) at wavelength 260 nm and 280 nm and the ratio of OD260nm/OD280nm was taken as a criterion to check the purity of DNA. The quality of the genomic DNA was checked by agarose gel electrophoresis (Bio-Rad Gel Doc™ EZ imager).
Genotyping
Agena MassARRAY™ platform was used for SNP genotyping, in Central Analyzer MassARRAY™ facility at SMVDU. It is a robust, cost-effective and highly sensitive tool for genotyping of SNPs and involves multiplex PCR [17]. Customized forward, reverse and single base extension primers were designed using Agena Design Suite V.2.0. Multiplex PCR was used to detect a variation in initially targeted region.1 µl of genomic DNA (concentration of 10 ng/ul) was loaded in 384 well PCR plates and dried at 85 °C for 10 min. After drying, the reaction mixture was prepared containing dNTPs, primers pool (forward & reverse), reaction buffer and DNA polymerase. After completion of first PCR, the reaction was treated with shrimp alkaline phosphatase (SAP). The multiplex PCR reaction was then subjected to single base extension reaction using mass modified ddNTPs and primers (pooled single extension primers). PCR cycle was adopted from Gabriel et al. 2009 [17]. Further the final PCR product is treated with cationic resin and then energy reaction to keep check the quality of genotyping and transferred to spectro-chip. The transferred product then fired to MT analyser. The data was then processed and analysed by preinstalled Typer Analyzer v.4.0. The genotyping results were recognized by replicating 10% of random samples and the concordance rate were 98.3%. In the reaction of 384 well plates one negative and one positive control were added to check the quality of reaction mixture.
Genotyping quality control
The accuracy and precision of downstream data analyses are highly dependent on the data quality of single nucleotide polymorphism (SNP) arrays. Low-quality genotypes can lead to false-positive results and reduce the precision of genomic predictions. Individual call rate, defined as the proportion of SNPs per individual where a genotype was called, is a common quality control measure [18]. So SNPs having call rate above 90% were included for statistical analysis [19]. Hardy–Weinberg Equilibrium (HWE) among cases and controls were used for assessing the quality of genotypes after analysis.
Statistical analysis
The statistical analysis was performed using Plink V.1.0962 with a maximum of 10,000 permutations [20]. Each SNP was subjected to Hardy Weinberg Equilibrium (H.W.E) and significant association of SNPs was evaluated by 3 × 2 chi square tests for genotypic frequencies between cases and controls. Further logistic regression analysis was performed using SPSS V.23 in order to obtain corrected odds ratio (OR), confidence interval (CI) and p value as level of significance from confounding factors like age and BMI. The power of the study was calculated using PS: power and sample size calculation (PS version 3.1.6) software and the power of study was > 90% (41).
Results
The current case–control association study included a total of 492 participants with 100 colorectal cases and 392 healthy controls. In the current study, the patient cohort included 56 males and 44 females, with a average age and BMI of 62.87 (± 9.8) years and 20.75 (± 0.869) kg/m2 respectively. Healthy controls constituted 280 males and 112 females with average age and average BMI of 48.81 (± 15.3) years and 24.9 (± 0.869) kg/m2 respectively. The clinicopathological characteristics of all the participants are shown in (Supplementary Table 1).
In the current study, genetic variants that were not studied in association with CRC in the population of Jammu and Kashmir but are associated with other types of cancer were evaluated. These genetic variations were studied to know whether they show increased risk or reduced risk with colorectal cancer in the population of Jammu and Kashmir. This is the first study to find the association of colorectal cancer in the population of Jammu and Kashmir.
We analysed 7 SNPs using MassARRAY™ After stringent quality check, these SNPs having genotyping quality call greater than 90% (Table 1).
This is the first study to explore these SNPs with colorectal cancer within the J&K population. Seven SNPs were selected for the study, out of these five SNPs rs2234593, rs1799966, rs2229080, rs8034191, rs1042522 were found to be significantly associated with the colorectal cancer under the allelic model with an odds ratio OR = 2.981(1.731–5.136 at 95% CI); p value = 4.81E-05 for rs2234593, OR = 1.685(1.073–2.647 at 95% CI);; p value = 0.02292 for rs1799966, OR = 1.5 (1.1–2.3 at 95% CI), p value = 0.02 for rs2229080, OR = 1.699(1.035–2.791 at 95% CI); p value = 0.03521 for rs8034191, OR = 20.07 (11.26–35.75); p value = 1.84E-34 for rs1042522 respectively.
Discussion
Colorectal cancer (CRC) is the 3rd most persistent cancer and a prominent cause of cancer-related morbidity and mortality worldwide [1]. CRC evolves due to the progressive accumulation of genetic and epigenetic modification in the colonic epithelium, transforming them into colorectal adenomas and adenocarcinomas [21].
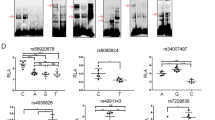
Genetics is an important risk factor associated with CRC. So, in the present study, genetic elucidation among cases and controls was explored. We analyzed seven genes in 492 samples consisting of 100 cases and 392 controls. All the seven SNPs had a call rate above 90%. These identified variants were rs2229080 of DCC, rs10046 of CYP19A1, rs1042522 of TP53, rs10228682 of POT1, rs10069690 of TERT, rs1051266 of SLC19A1, and rs1026071 of ARTNL (Supplementry Fig. 1). GWAS threshold is a statistical threshold for establishing the statistical significance of a claimed relationship between a single-nucleotide polymorphism (SNP) and a characteristic in genome-wide association studies. The most widely used threshold is p < 5 × 10 − 8, which is calculated by applying a Bonferroni adjustment to all of the independent common SNPs across the the human genome. Table 1 shows that five out of seven SNPs were in a strong association; however, rs1042522 of TP53 have GWAS threshold value.And to check the Putative Role of the associated variants with Colorectal we used GTEx Table 2
rs2234593 of WT1 (Wilms tumor 1, transcription factor)
This tumor suppressor gene plays an important role in cell growth and apoptosis. The chromosomal location of WT1 is 11 and it has been found to be associated with acute myeloid leukemia, lung cancer, brain tumors, breast cancer, colorectal adenocarcinoma, thyroid cancer, desmoid tumors, etc. [22, 23]. In a study it was found that the variant rs2234593 of WT1 was linked with overall survival (OS) and relapse in cancer (leukemia) patients[24]. Another study signified overexpression of WT1 in colorectal cancer[25]. In the present study variant rs2234593 of WT1 has shown no association with CRC in the J&K population.
rs1799966 of BRCA1 (breast cancer type 1 susceptibility protein)
The variant rs1799966 of BRCA1 has been proved to be linked with “pancreatic cancer” with a “hazard ratio” of 1.23 (95% CI: 1.09–1.40, P = 0.0010) in the population of China [26] but not associated with possibility of breast carcinoma [27]. It has been confirmed that mutations in BRCA1 or BRCA2 results in multiple cancers like colorectal cancer and pancreatic adeno-carcinoma [28, 29]. In present study, the genetic variant rs1799966 of BRCA1 has been evaluated with respect to colorectal cancer and it was observed that the variant under study was found to be associated with the higher risk of colorectal cancer in the J&K population with O.R 1.685 (1.073 to 2.647, at 95%CI, p value = 0.022).
rs2229080 of DCC (“deleted in colorectal carcinoma, netrin 1 receptor”)
DCC (netrin-1), initially discovered in CRC, encodes the netrin-1 receptor, a member of the cell's immunoglobulin superfamily adhesion molecules, has been characterized as a potential tumor suppressor gene [30,31,32]. As soon as DCC bind to the netrin-1 receptor, it induces cell migration and proliferation. In the absence of netrin-1, DCC's intracellular domain is cleaved by a caspase that induces apoptosis in a caspase-9-dependent pathway (Supplementry Fig. 2) [33]. DCC is frequently silenced or inactivated in various human cancers due to epigenetic silencing or loss of heterozygosity at chromosome 18q21 region [31, 34]. Loss of DCC gene expression was shown to be an independent prognostic factor in colorectal [12], AML [35], and gastric cancer [13, 36] patients. Various studies have been carried out that demonstrate a significant association of DCC polymorphism with esophageal, colorectal, and gastric cancer risk [16, 37,38,39]. Further, rs2229080, a missense variation replacing Arg to Gly at DCC codon 201, was reported to increase the risk of colorectal cancer [40] and neuroblastoma [41]. In present study, the genetic variant rs2229080 of DCC has been evaluated with respect to colorectal cancer and it was observed that the variant under study was found to be associated with the higher risk of colorectal cancer in the J&K population with OR = 1.5 (1.1–2.3 at 95% CI), p value = 0.02.
rs1801133 of MTHFR (methylenetetrahydrofolate reductase)
An enzyme called “methylenetetrahydrofolate reductase” is produced with the instructions from the “MTHFR”. This enzyme has a function in the processing of amino acids, the basic components of proteins [42]. Although a number of mutations were described, 1298A > C (rs1801131) and 677C > T (rs1801133) “single nucleotide polymorphisms” (SNPs) are the two most general mutations in the MTHFR [43]. For the creation of a thermo labile variety of MTHFR, these two identified polymorphisms are responsible [43]. There was a common presence of the 677 TT genotype in southern Italy (26%), Mexico (32%) and Northern China (20%). The 677C > T mutation “rs1801133” in the “MTHFR” is a key reason for mild “hyperhomocysteinemia”, while at nucleotide position 1298, the second polymorphism is not so well described [44]. However, a meta-analysis by Zan Teng (2013) suggests that the possibility of getting “colorectal cancer”increases with the MTHFR variant rs1801133 polymorphism (677C > T), whereas there is no link between African people in the “subgroup analysis by ethnicity” [45]. In this research, we tried to find the relationship of the variant rs1801133 of MTHFR with CRC among the J&K population but the differences in the allelic frequency distribution of rs1801133 variant between cases and controls were statistically insignificant.
rs10046 of CYP19A1 “cytochrome P450 family 19 subfamily a member 1”
Many researches has stated that the rs10046 variant of CYP19A1 is linked with gastric,breast, lung, and colorectal cancer [46, 47]. The current research tried to explore the association of variant rs10046 with CRC in Jammu & Kashmir but the differences in the allelic frequency distribution of rs10046 variant between cases and controls were statistically insignificant.
rs8034191 of HYKK (hydroxylysine kinase)
HYKK is a protein coding gene located at chromosome 15. The HYKK was reported to be associated with the susceptibility of lung cancer [48, 49]. Many studies has signified that the variant rs8034191 of HYKK is linked with “lung cancer” possibility [49,50,51,52] but not linked with colorectal cancer worldwide. There were no studies found in the database that describes about the function of the variant rs8034191 of HYKK in CRC in the Indian population. In the present study, the variant rs8034191 of HYKK has been evaluated with the colorectal cancer in J&K region of India. The results of the study reported that variant rs8034191 of HYKK was associated with the higher risk of colorectal cancer in Jammu and Kashmir population with O.R 1.699 (1.035 to 2.791, at 95% CI, p = 0.035). This is the first study that has investigated and reported significant association of variant rs8034191 of HYKK with colorectal cancer in the population of Jammu and Kashmir, India.
rs1042522 off TP53 (tumor protein p53)
The “TP53”gives instructions for the production of a protein known as p53 tumor protein. This gene encodes a “tumor suppressor protein” that includes domains of DNA binding, oligomerizationand transcriptional activation [53]. Due to its function in inhibiting cancer growth and regulating cell division, p53 is also called the “guardian of the genome”. TP53 mutations are universal across several cancer types [54]. The loss of a tumor suppressor is most often caused by important harmful events, such as frame change mutations or premature stop codons. Abnormalities of the tumor suppressor gene, such as those of TP53, are common but are currently not clinically actionable [55]. Many studies have indicated that the “TP53” (rs1042522 C > G) polymorphism is linked with susceptibility to differentforms of cancer like cervical cancer, breast cancer, lung cancer, CRC, endometrial cancer, and ovarian cancer [56, 57]. While a retrospective study in Taiwan region, it was found that the carriers of the “C allele” of variant rs1042522 were linked with a reduced colorectal cancer risk [58]. In present study, the genetic variant rs1042522 of TP53 has been evaluated with respect to colorectal cancer and it was observed that the variant under study was found to be associated with the higher risk of colorectal cancer in the J&K population with OR = 20.07 (11.26–35.75); p value = 1.84E-34.
To the best of our knowledge, to date, no study has been conducted on the role of the Single Nucleotide variants rs2234593, rs1799966, rs2229080, rs8034191, and rs1042522 in colorectal cancer within the population of J&K. This is the first prelude study that investigated the possible correlation between the rs2234593, rs1799966, rs2229080, rs8034191, and rs1042522 polymorphisms and susceptibility to colorectal cancer.
Conclusion
In the present study, we explored the link between environmental factors, genetics, and colorectal cancer. This study is the first to investigate the relation of genetic variants associated with colorectal cancer within Jammu and Kashmir. The present study could provide insights into genetic variation associated with the risk of developing colorectal cancer. Hence, if investigated further in the large cohort, this can unravel the biological significance of these SNPs in colorectal cancer among the Jammu and Kashmir populations.
Data availability
Data generated and analyzed during study is not available publicly but can be made available from the corresponding author upon reasonable request.
References
Sung H et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71(3):209–249
Rawla P, Sunkara T, Barsouk A (2019) Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Gastroenterol Rev 14(2):89
Baena R, Salinas PJM (2015) Diet and colorectal cancer. Maturitas 80(3):258–264
Jung G et al (2020) Epigenetics of colorectal cancer: biomarker and therapeutic potential. Nature Rev Gastroenterol Hepatol 17(2):111–130
Bogaert J, Prenen H (2014) Molecular genetics of colorectal cancer. Ann Gastroenterol 27(1):9
al., K.U.N.e., Survey of Patients with Cancer in Jammu and Kashmir: Based on Hospital Registry Records. Indian Journal of Computer Science and Engineering (IJCSE), 2017.
Giovannucci E et al (1995) Physical activity, obesity, and risk for colon cancer and adenoma in men. Ann Intern Med 122(5):327–334
Shah R et al (2020) MassARRAY analysis of twelve cancer related SNPs in esophageal squamous cell carcinoma in J&K, India. BMC Cancer 20(1):1–6
Verma S et al (2020) Genetic variants of DNAH11 and LRFN2 genes and their association with ovarian and breast cancer. Int J Gynaecol Obstet 148(1):118
Bhat A et al (2019) Association of ARID5B and IKZF1 variants with leukemia from Northern India. Genet Test Mol Biomarkers 23(3):176–179
Ellis JA, Ong B (2017) The MassARRAY® system for targeted SNP genotyping. Genotyping. Springer, New York, pp 77–94
Sun XF et al (1999) Expression of the deleted in colorectal cancer gene is related to prognosis in DNA diploid and low proliferative colorectal adenocarcinoma. J Clin Oncol 17(6):1745–1750
Yoshida Y et al (1998) Decreased DCC mRNA expression in human gastric cancers is clinicopathologically significant. Int J Cancer 79(6):634–639
Onwe EE et al (2020) Predictive potential of PD-L1, TYMS, and DCC expressions in treatment outcome of colorectal carcinoma. Cancer biology and advances in treatment. Springer, Cham, pp 97–112
Liu X et al (2016) Genetic association of deleted in colorectal carcinoma variants with breast cancer risk: a case-control study. Oncotarget 7(22):32765–32773
Malik MA et al (2013) Role of genetic variants of deleted in colorectal carcinoma (DCC) polymorphisms and esophageal and gastric cancers risk in Kashmir Valley and meta-analysis. Tumour Biol 34(5):3049–3057
Gabriel S, Ziaugra L, Tabbaa D (2009) SNP genotyping using the sequenom massARRAY iPLEX platform. Curr Protoc Hum Genet 60(1):2.12.1-2.12.18
Purfield DC, McClure MC, Berry DP (2016) 0295 The impact of call rate on genotype accuracy. J Anim Sci 94(suppl_5):140–141
Anderson CA et al (2010) Data quality control in genetic case-control association studies. Nat Protoc 5(9):1564–1573
Purcell S et al (2007) PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81(3):559–575
Jung G et al (2020) Epigenetics of colorectal cancer: biomarker and therapeutic potential. Nat Rev Gastroenterol Hepatol 17(2):111–130
Oji Y et al (2002) Overexpression of the Wilms’ tumor gene WT1 in de novo lung cancers. Int J Cancer 100(3):297–303
Silberstein GB et al (1997) Altered expression of the WT1 Wilms tumor suppressor gene in human breast cancer. Proc Natl Acad Sci USA 94(15):8132–8137
Niavarani A et al (2016) The Wilms tumor-1 (WT1) rs2234593 variant is a prognostic factor in normal karyotype acute myeloid leukemia. Ann Hematol 95(2):179–190
Oji Y et al (2003) Overexpression of the Wilms’ tumor gene WT1 in colorectal adenocarcinoma. Cancer Sci 94(8):712–717
Zhu Y et al (2017) BRCA1 missense polymorphisms are associated with poor prognosis of pancreatic cancer patients in a Chinese population. Oncotarget 8(22):36033
Yang M et al (2019) Association between BRCA1 polymorphisms rs799917 and rs1799966 and breast cancer risk: a meta-analysis. J Int Med Res 47(4):1409–1416
Katona BW et al (2019) RE: BRCA1 and BRCA2 gene mutations and colorectal cancer risk: systematic review and meta-analysis. J Natl Cancer Inst 111(5):522–523
Mersch J et al (2015) Cancers associated with BRCA 1 and BRCA 2 mutations other than breast and ovarian. Cancer 121(2):269–275
Rai R et al (2015) A multiple interaction analysis reveals ADRB3 as a potential candidate for gallbladder cancer predisposition via a complex interaction with other candidate gene variations. Int J Mol Sci 16(12):28038–28049
Fearon E et al (1990) Identification of a chromosome 18q gene that is altered in colorectal cancers. Science 247(4938):49–56
Rai R et al (2013) DCC (deleted in colorectal carcinoma) gene variants confer increased susceptibility to gallbladder cancer (Ref. No.: gene-D-12–01446). Gene 518(2):303–9
Forcet C et al (2001) The dependence receptor DCC (deleted in colorectal cancer) defines an alternative mechanism for caspase activation. Proc Natl Acad Sci 98(6):3416–3421
Arakawa H (2004) Netrin-1 and its receptors in tumorigenesis. Nat Rev Cancer 4(12):978–987
Inokuchi K et al (2002) Loss of DCC gene expression is of prognostic importance in acute myelogenous leukemia. Clin Cancer Res 8(6):1882–1888
Bamias AT et al (2003) Prognostic significance of the deleted in colorectal cancer gene protein expression in high-risk resected gastric carcinoma. Cancer Invest 21(3):333–340
Starinsky S et al (2005) Genotype phenotype correlations in Israeli colorectal cancer patients. Int J Cancer 114(1):58–73
Djansugurova L et al (2015) Association of DCC, MLH1, GSTT1, GSTM1, and TP53 gene polymorphisms with colorectal cancer in Kazakhstan. Tumor Biol 36(1):279–289
Toma M et al (2009) P53 and DCC polymorphisms and the risk for colorectal cancer in Romanian patients—a preliminary study. J Analele Univ Oradea Fasc Biol 16:162–165
Minami R et al (1997) Codon 201Arg/Gly Polymorphism of DCC (deleted in colorectal carcinoma) gene in flat- and polypoid-type colorectal tumors. Dig Dis Sci 42(12):2446–2452
Kong X-T et al (2001) Codon 201Gly polymorphic type of the DCC gene is related to disseminated neuroblastoma. Neoplasia 3(4):267–272
Fenwick R (2016) The eyes never lie gene: explanation of the Mthfr 677 C / A cytogenic location 1p 363 short (P) arm of chromosome 1. Int J Complement Altern Med. https://doi.org/10.15406/ijcam.2016.03.00101
Nefic H, Mackic-Djurovic M, Eminovic I (2018) The frequency of the 677C> T and 1298A> C polymorphisms in the methylenetetrahydrofolate reductase (MTHFR) gene in the population. Med Arch 72(3):164
Cortese C, Motti C (2001) MTHFR gene polymorphism, homocysteine and cardiovascular disease. Public Health Nutr 4(2b):493–497
Teng Z et al (2013) The 677C> T (rs1801133) polymorphism in the MTHFR gene contributes to colorectal cancer risk: a meta-analysis based on 71 research studies. PLoS ONE 8(2):e55332
Friesenhengst A et al (2018) Elevated aromatase (CYP19A1) expression is associated with a poor survival of patients with estrogen receptor positive breast cancer. Horm Cancer 9(2):128–138
Artigalás O et al (2015) Influence of CYP19A1 polymorphisms on the treatment of breast cancer with aromatase inhibitors: a systematic review and meta-analysis. BMC Med 13(1):1–10
Wang J et al (2017) Genetic predisposition to lung cancer: comprehensive literature integration, meta-analysis, and multiple evidence assessment of candidate-gene association studies. Sci Rep 7(1):1–13
Weissfeld JL et al (2015) Lung cancer risk prediction using common SNPs located in GWAS-identified susceptibility regions. J Thorac Oncol 10(11):1538–1545
Ji X et al (2018) Identification of susceptibility pathways for the role of chromosome 15q25.1 in modifying lung cancer risk. Nature Commun 9(1):3221
Hu B et al (2014) Quantitative assessment of the influence of common variations (rs8034191 and rs1051730) at 15q25 and lung cancer risk. Tumor Biol 35(3):2777–2785
Gu M et al (2012) Strong association between two polymorphisms on 15q25. 1 and lung cancer risk: a meta-analysis. PLoS ONE 7(6):e37970
Sieren JC et al (2014) Development and translational imaging of a TP53 porcine tumorigenesis model. J Clin Invest 124(9):4052–4066
Giordano TJ (2014) The cancer genome atlas research network: a sight to behold. Endocr Pathol 25(4):362–365
Takahashi T et al (1989) p53: A frequent target for genetic abnormalities in lung cancer. Sci 246(4929):491–494
Ahmed N, Abubaker K, Findlay JK (2014) Ovarian cancer stem cells: molecular concepts and relevance as therapeutic targets. Mole Asp Med 39:110–125
Klug SJ et al (2009) TP53 codon 72 polymorphism and cervical cancer: a pooled analysis of individual data from 49 studies. Lancet Oncol 10(8):772–784
Kamiza AB et al (2016) TP53 polymorphisms and colorectal cancer risk in patients with Lynch syndrome in Taiwan: a retrospective cohort study. PLOS ONE 11(12):e0167354
Acknowledgements
All authors acknowledge all the participants for their contribution in this study and. BS acknowledges Dr. Varun Sharma and Dr. Indo Sharma for their valuable suggestions.
Funding
No Funding.
Author information
Authors and Affiliations
Contributions
RS and RK planned the work. BS, AB and SV performed experimental work in lab. GR, AB, RJ, SV, AA and DB helped in sample collection. SA gave the histopathologically conformation of the samples. BS performed the statistical analysis, BS and RJ wrote the manuscript and RJ drafted the manuscript according to journal. RS, RK, and RA finally refined the manuscript. All authors finally revised and approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interest.
Ethical approval
The study was approved by the Institutional Ethics Review board (IERB) of Shri Mata Vaishno Devi University (SMVDU). In this study all experimental research work was performed according to guidelines issued by Institutional Ethics Review board committee.
Consent to participate
The written informed consent was obtained from each participant before conducting the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been already submitted to a preprint platform, DOI: 10.21203/rs.3.rs-199948/v1, License: CC BY 4.0.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sharma, B., Angurana, S., Bhat, A. et al. Genetic analysis of colorectal carcinoma using high throughput single nucleotide polymorphism genotyping technique within the population of Jammu and Kashmir. Mol Biol Rep 48, 5889–5895 (2021). https://doi.org/10.1007/s11033-021-06583-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06583-8