Abstract
Forensic DNA typing and subsequent molecular methods of sex determination in humans have been proven to be an imperious tool to criminal justice system. In current practice, most of the short tandem repeat (STR) based commercial kits contain amelogenin as the sexing marker. Amelogenin gene which contributes to the tooth enamel formation is present on both X and Y chromosome with a variation in base pair size. However, huge discrepancies have been observed with amelogenin based sex determination mostly due to X and Y deletion in the population and mutation in primer binding sites. Some ethnicities such as those in Indian population are affected badly with inappropriate sex determination by amelogenin marker due to the presence of high frequency of Y deletion in the population. Presence of PCR inhibitors, degradation in the DNA samples and presence of mixed DNA also contribute to the discrepancy in results obtained by amelogenin analysis. To overcome this problem, many alternative markers/techniques such as STS, SRY, TSPY, DXYS156, SNPs, DYZ1 and Next generation sequencing have been discussed in much detail with their respective pros and cons. In this regard, inclusion of one or more alternative markers along with amelogenin will decrease the anomalies in sex determination observed while using the amelogenin marker alone in forensic sample analysis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
DNA fingerprinting has become an imperative tool in forensic science and provides a valuable aid to the criminal justice system. The current trend of forensic DNA typing relies on the analysis of a combination of short tandem repeats (STR) markers from the DNA isolated from the questioned biological samples for its possible match with the reference samples with highest degree of fidelity [16]. Molecular analysis through DNA typing is not only useful for unravelling criminal and civil cases of varied nature such as paternity disputes, identification of mass disaster victims, investigations of missing persons and sexual assault cases, but also determines the correct genetic sex of an individual [89]. Though many phenotypic parameters have been reported for the sex determination of a skeletonized body, molecular methods of sex determination have been proven to be superior to any other currently available technique [14]. Additionally, most of the phenotypic tests such as cranial and pelvic traits, meatus acusticus internus characteristics, anthropometric measurements of the limbs and hands require intact bone(s), which are mostly unavailable in some forensic scenarios [1].
In this regard, Y chromosome specific markers are highly useful in sex determination of human skeletal remains as well as the origin of the biological samples [62]. These markers can also provide vital information in sexual assault cases to determine the presence of male DNA in the victim’s belongings. Most of the currently available commercial forensic DNA typing kits have included amelogenin marker for sex determination along with other STR markers [122]. US National DNA Index System (NDIS) and Combined DNA Index System (CODIS) have also recommended Amelogenin marker to be included for sex determination during forensic DNA typing analysis [45]. However, discrepancies in the results of amelogenin based sex determination are widely reported throughout the globe [73]. Failure of amelogenin marker for sex determination has been attributed to many factors, such as genetic basis of ethnicity, degradation of biological sample, presence of PCR inhibitors and mutations in primer binding sites to mention a few.
In forensic cases, the molecular technique to be used for sex determination should produce highly precise, reliable and consistent results and any chance of possible errors should be seconded by additional/alternative techniques. To overcome this problem, many Y specific markers for sex determination are flourishing day by day to be used either independently or in combination with amelogenin to produce highly reliable results. Some of the possible alternative markers include the STS gene, SRY gene, TSPY, DXY156, DYZ1 and many SNPs [15]. Additionally, advanced techniques such as NGS and pyrosequencing have also proved to be useful in sex determination at molecular level. In this regard, this article discusses the possible alternative markers and their expediency in distinguishing sex with higher fidelity in forensically relevant samples.
Amelogenin: currently used exclusive sex determination marker in forensic application
Amelogenin gene produces a heterogeneous population of mRNAs due to alternative splicing, subsequently generating heterogenous group of amelogenin proteins responsible for tooth enamel matrix formation [49]. Though other genes and/or proteins are also involved in tooth enamel formation, 80–90% of the same is formed by the amelogenin protein [36] (Fig. 1). Thus, any mutation/alteration in the amelogenin locus cause a disease affecting the structure and appearance of enamel, called amelogenesis imperfect [103]. AMELX is the main source of such mutations and this disease has not yet been connected with any mutations in the AMELY. Various studies have concluded that X linked amelogenesis imperfecta is a heterogeneous disease having different phenotypic effects in varied cases [66]. A deletion of over 5 kb in the amelogenin has been found to cause amelogenesis imperfecta in males while in females, the cause has been found to be a non-sense mutation in exon 5 [25]. Likewise, a recent study has identified a deletion in the Amelotin (AMTN) which is a matrix protein responsible for mineralization during enamel development, responsible for Amelogenesis Imperfecta [108].
In humans, both AMELX and AMELY genes are present on sex chromosomes at p22.1-22.3 region and at Yp11.2 region respectively, which are responsible for synthesizing the same Amelogenin protein [91]. AMELX is responsible for 90% of the amelogenin proteins whereas only 10% is contributed by AMELY [7]. Nucleotide sequences show 89% homology between AMELX and AMELY along with the presence of highly conserved protein coding regions at 2–6 exons [15]. Hence, the homology, location of the genetic marker and distinguishability between AMELX and AMELY make it a suitable marker for genetic basis of sex determination in forensic DNA typing applications.
Usefulness of amelogenin marker in sex determination
Amelogenin based sex determination relies on the presence of two homologues of Amelogenin i.e. AMELX and AMELY, which exhibit length polymorphism. They differ both in size and sequence and hence can provide sex differentiation. In comparison to AMELY, AMELX possesses a 6 bp deletion contributing to the difference in size between these two homologous genes. PCR amplification using the commonly used primer sets produce fragments of 106 and 112 bp in length for AMELX and AMELY respectively [82]. Thus, in females only X specific fragment can be observed, whereas, in males both X and Y specific fragments are observed after electrophoretic separation [33]. Hence, the male and female can be distinguished on the basis of the number of fragments detected in them [91, 103].
Sullivan et al. [110] initially developed a single set of primers spanning part of the intron 1 of Amelogenin gene for quantitative assessment of sex at DNA level using less than 1 ng DNA template. Gradually, to accommodate amelogenin marker in multiplexing systems, varied primer sets have been developed targeting different fragments of the gene and flanking regions. In this context, intron 3 of the Amelogenin gene has been explored greatly to design primer sets targeting the 6 bp difference in amplicons between AMELX and AMELY (Table 1).
Discrepancies in amelogenin based sex determination
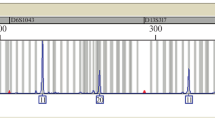
Amelogenin has been used for decades for sex determination at molecular level in forensics as well as for genetic and molecular profiling in archeology and anthropology. In non-human studies, amelogenin marker is useful in the reproduction field, archaeozoology, meat production and sample identification [37]. It is included in all the present day available genotyping kits and no other substitute has ever been proposed. The efficacy of the amelogenin marker came into question when cases of false detection of males as females started surfacing [128]. The amelogenin failure cases and their studies have increased several folds in the last decade. Amelogenin marker may give false negative results due to allele dropouts resulting from either deletions or mutations in the same region. Males harboring deletions and mutations in the AMELY region would be identified as females in the absence of any other corroborating evidences. Discrepancies in the Amelogenin based sex determination due to allele dropouts can result in different false positive and false negative results as explained in Fig. 2. Additionally, deletions in the amelogenin region of the Y chromosome are far more frequent than in the X chromosome [73, 83, 109]. The failure due to deletions can range up to 10% in an endogamous population. However, such discrepancies can be detected by sequencing and using different genotyping kits or different sets of primers [60, 77, 85]. The different nature of genetic anomalies resulting in the non-coherent discrimination between males and females has been described in details.
Mutation in the primer binding region
One of the major causes of allele dropout is mutation in the primer binding region of the amelogenin gene. Failure in primer binding results in allele dropout which ultimately results in misidentification of the gender. Such mutations prevent the binding of primer and hence the amplification of AMELY [101, 106]. As a result, a male can be falsely identified as a female due to the absence of AMELY fragment in the amplified product. A study by Borovko et al. [13] reported two cases of AMELX and AMELY dropout due to mutations in primer binding sites in Belarusian population. Another study involving 5534 subjects of Polish population showed the failure of AMELX amplification in 0.02% of cases using SGMPlus, ProfilerPlus and PowerPlex16 kits. However, when alternative primer sets are used, an X homologue product was amplified confirming the mutation in primer binding site [78]. Further studies showed that mutations may cause failure in some of the cases but most failures are caused by a deletion in the AMELY region [77, 109].
Y deletion
Most of the cases of amelogenin failure result from the absence of AMELY in the genotyping results when the subject had the Y chromosome. These discrepancies are due to deletion of nucleotide sequences on the Y chromosome that includes the AMELY region [71]. In cases of Y deletion, known male samples show only the presence of AMELX with 6 bp deletion resulting in the appearance of only one peak after genotyping. In these cases, the presence of Y chromosome can be assured either by karyotyping or by the use of alternative markers present on Y chromosome [83]. Such Y deletions have been reported on 6 instances by a study involving 29,432 male individuals of Australian population [109]. Other study involving an Indian population confirmed Y deletion at a higher rate in Indian population appearing in five cases when 270 individuals were subjected to examination [113]. Y deletion is present in almost all populations throughout the globe and their corresponding frequencies of occurrence have been given in Table 2. As major part of Y chromosome does not undergo recombination, any anomalies in the Y chromosome are passed as such to the next generations leading to the formation of geographically specific haplogroups [97]. In this context, haplogroups belonging to the Indian and Asian Ancestry have been found to have a higher frequency of Y deletion.
Y deletion mapping
Deletions in the AMELY region are usually studied by sequencing upstream and downstream markers surrounding the AMELY region. A combination of sequence-tagged-site (STS) deletion mapping, binary-marker, Y-STR haplotyping and TSPY copy number estimation is used to study these deletions by referring to the reference database sequence of the Y chromosome [57]. Further, the size of the deletions can be detected on the basis of presence of desired markers in the amplified product [60]. Many studies have reported the size of deletion to be approximately 2.5 Mb [71, 112]. Primers devised in earlier studies along with new set of primers, are generally used for Y deletion mapping. Using different set of primers leads to generation of different size of deleted products. Jobling et al. [57] mapped the deletions by designing Y specific STSs and verifying STSs near the AMELY region for Y specificity in already published STSs. Additionally, Mitchell et al. [85] estimated the size of Y deletion in 5 AMELY null males of Australian population to be of two different sizes, ranging from 304 to 731 Kbp and 712 to 1001 Kbp. For this study, markers surrounding the AMEL locus were targeted to map the size of deletions and confirmed the presence of the deletions on the short arm of the Y chromosome. A further study by Jobling et al. [57] speculated the presence of five different classes of Y deletion after examining 33 Y STR markers i.e. Class I, I-sY59, II, III and IV. However, study of Takayama et al. [112] reported a Y deletion that did not belong to any of the above mentioned classes. Though AMELY deletion reveals a normal tooth phenotype, it has many classes. In Class I deletion (known as PRKY), along with AMELY, serine-threonine protein kinase and TBL1Y normally expressed in prostate and foetal brain also get deleted. Whereas, in non-Class I deletions, Y linked PCDH11Y (Protocadherin 11) expressed in brain is lacking. Another reported Class III case lacked testis specific Y linked Transforming Growth Factor-Beta-Induced Factor 2-Like (TGIF2LY) [57].
AMELY deletion in Indian population
The failure rate of amelogenin varies among populations throughout the globe. However, the maximum failure rate has been observed in the Indian subcontinent populations. The landmark studies regarding the Y deletion in Indian population have been reported by Santos et al. [102], Kashyap et al. [60] and Thangaraj et al. [113]. Various studies reporting frequencies of Y deletions in Indian as well as other populations have been given in Table 2. Human Y chromosome haplogroup represents the mutations in the non-recombining portions of the Y chromosome. This represents the common paternal descends [69]. About 5% of Indian males belong to, the J2 haplotype lineage harboring mutation at M172 genetic marker [6]. This lineage originated in the Middle East and was later distributed across Europe and Asia. Indian males belonging to the J2 lineage are found to have more frequent Y deletions [57, 60, 77, 127]. Additionally, “Haplogroup O” of Indian population has been found to possess a 5 bp deletion at position M175, whereas 1 bp deletion is common at M17 UEP in M173 lineage [130]. In this context, Kashyap et al. [60] concluded that Y deletion in amelogenin is lineage specific and inherited from one generation to another. Similarly, Cadenas et al. [18] suggested a common ancestry for AMELY null males on the basis of a finding that reported 5 AMELY null males belonging to the same lineage i.e. J2b2-M241. In this case, the Y STR profiles of two individuals were identical in nature whereas, other studied profiles had striking similarity.
X deletion
AMELX dropout is a rare phenomenon as compared to AMELY dropout. Mutation in AMELX can be the cause of AMELX dropout in males but it would not affect the gender identification as there would be normal amplification of AMELY. Additionally, in case of females, AMELX dropout is difficult to detect as it is very rare to have a mutation in both the homologous chromosomes and in cases of mutation in single chromosome, a single peak would be observed similar to a normal female as has been shown in Fig. 2. A study on the South Chinese population showed deletion of AMELX at a frequency of 0.037% [93]. Similarly, a female patient with clinical symptoms of microphthalmia has been reported to possess deletion of the entire AMELX gene along with additional 50 monosomic genes localized to Xp22 [47]. Two cases of AMELX dropout have also been identified in Belarusian population; however these have been attributed to the mutation in primer binding sites [13]. Use of Powerplex 16 System (Promega) has also been reported to generate only Y homologue of amelogenin in 10 male individuals of West African population [4].
Most of the currently used human STR kits such as, InnoTyper21 kit, AmpFLSTR™ Identifiler™ kit, PowerPlex® 21 System do not possess any alternative sex determining markers. However, few new generation kits such as GlobalFiler™ (Thermo Scientific), PowerPlex® Fusion 6C (Promega Crop.), Investigator 24plex GO! Kit (Qiagen) and SureID® PanGlobal (Health Gene Tech.) harbor DYS391, DYS570, DYS576 and Y InDel as alternative sex determining markers [30]. However, when AMELY deletion occurs, simultaneous deletion of DYS570 and DYS576 occurs as they are present within the AMELY region on the Y chromosome (Fig. 3) [59]. Additionally, the position of DYS391 is very close to the AMELY region, which increases the chance of co-deletion of DYS391 along with other Y-STR markers. Besides, none of the markers get amplified in female samples, thus becomes difficult to distinguish between the causes of non-amplification, i.e. female sample or sample with PCR inhibitors. Thus, alternative markers should be assessed to the currently available set of commonly used STR markers for sex determination.

Modified from Kareem et al. [59]
Relative positions of the commonly used Y-STR markers on the Y chromosome.
Amelogenin marker and PCR inhibitors
Sex determination using STR markers can be affected by PCR inhibitors, especially in cases of forensic samples. Commonly known PCR inhibitors may arise from the environmental sources, as microbial by-products or chemical contamination during DNA isolation. Though the mechanism of action among different PCR inhibitors differs, the PCR process being an enzymatic reaction is highly sensitive to them. These inhibitors can have profound effect on STR amplification and often result in dropouts, peak imbalance and stutters. Though no specific study on this aspect has been conducted till date, the potential role of these inhibitors in affecting the amplification process of amelogenin marker cannot be ignored. In this regard, a list of potential inhibitors affecting the Amelogenin amplification as well as other markers has been given by Dash and Das [26].
Degraded vs. mixed samples
Analysis of Amelogenin marker may cause ambiguity between degraded and mixed forensic samples. In case of mixed samples of two heterosexual origins, Amelogenin marker generates both X and Y peaks with imbalanced peak heights and a difference of 70% in magnitude [41]. On similar lines, degraded forensic samples also produce DNA profiles with heterozygous imbalance complicating the analysis process [44]. Most of the primer sets used for sexing using amelogenin marker require a minimum of 106 bp of template DNA for proper amplification. Many studies have reported high fragmentation of forensic samples which in turn generate DNA fragments as low as 61 bp [27, 76]. In this context, Fazi et al. [34] reported the limitation of amelogenin marker in as many as 28 degraded DNA samples from analyzed touch DNA and hair shaft sources. Limitations of using amelogenin marker in ancient DNA analysis have also been reported [82].
Alternative markers for sex determination
For forensic application, the molecular technique used should be fool proof and the technique(s) with even little chance of error should be seconded by additional or alternative techniques. Amelogenin may lead to various kinds of discrepancies in sex determination due to deletion and/or mutations depending on the nature and type of biological evidence. In this regard, a multiplex method involving other sex determination markers along with the amelogenin is the best proposed method for reliable sex determination to avoid any possible anomalies. Some of the alternative sexing markers with potential forensic applications have been described in detail. An extensive list of primer sets for amplification of the alternative sexing markers has been provided in Table 3. However, huge challenges come into existence when samples of forensic relevance are being analyzed. Hence, a comprehensive list of the alternative markers with their corresponding pros and cons has been given in Table 4.
Steroid sulfatase (STS)
Steroid sulfatase is a widely dispersed enzyme that plays an important role in the formation of steroids encoded by the STS gene found at the Xp22.3 [116]. It is pseudo autosomal in nature and on Y chromosome, a homologue of STS is present as a pseudogene which is transcriptionally inactive. Certain point mutations and/or deletions have been reported in this gene resulting in the development of X linked Ichthyosis as well as other neurodevelopmental disorders like Attention-deficit hyperactivity disorder (ADHD) and Autism [42].
STS-X, sized about 146 kb with intron 1 of 35 kb is the prime target for development of sex determination marker [126]. Morikawa et al. [86] developed primers for amplifying the first intron of the STS gene and its homologue on the Y chromosome resulting in the DNA fragments of 158 and 166 bp on X and Y chromosome respectively. This size variation between the amplicons is used to differentiate STS and its homologue on Y chromosome i.e. STSP1 and hence can be employed for sex determination [86]. STS sequences have been reported successful for sex determination in 53 mammalian species with greater efficacy [48]. Additionally, Morikawa et al. [86] used certain additional markers including STS to confirm the sex of an individual to be male, which was incorrectly reported to be female by the amelogenin analysis.
SRY gene
The SRY gene, also known as the sex determining region is responsible for the determination of male sex in humans as it promotes the development of precursor cells into sertoli cells instead of follicle cells [43]. SRY is a transcriptional regulator that directly up-regulates the SOX9 i.e. Sry box9. It dominates the Wnt/b-catenin pathway which promotes the female development and in turn allows expression of SOX9 to initiate male sex development [10]. SRY acts only on the sex gonads and the rest of the body is conveyed the sex determination message via hormones i.e. if the male gonads are removed in fetal stage, the embryos with XY phenotype would develop as female [80]. Additionally, mutation in SOX9 may generate a female individual by affecting the development of Y chromosome [54]. Being directly related to the male phenotype, this marker has an edge over others.
A new set of primers has been developed by Drobnič [32] for determining sex using SRY marker. Similarly, Steinlechner el al. [109] successfully depicted the use of SRY marker for sex determination in males with amelogenin deletion. A similar study on forensic samples confirmed the sensitivity and reliability of this marker for forensic applications either as a single marker or in multiplex system with high efficacy in mixture analysis [61]. In this regard, Codina et al. [24] developed a multiplex system involving the simultaneous amplification of amelogenin, SRY and four mini X-STR loci for application in sex determination of mixed as well as degraded forensic samples. Additionally, SRY marker has also shown its potential advantage over other sexing markers in sex determination of challenged forensic samples i.e. incinerated teeth [90].
TSPY
TSPY is a multi-copy gene family with repetitive units and a variation of copy number in the range of 30-60 bp located on the short arm of Y chromosome and regulates the process of spermatogenesis [124]. The whole gene consists of six exons and five introns which approximate to about 2.8 kb in size. A recent study has concluded that, men with less than 33 copies of TSPY have a 1.5 times higher risk of infertility [39, 119]. A minimum of 6 locations on the Y chromosome have been reported to be occupied by TSPY gene family members [28].
As several genes of the TSPY are present on the Y chromosome, different primers can be designed to exhaust the high repeats of this marker for sex determination. As TSPY is only present on the Y chromosome, female samples will show no peak in the electropherogram for the TSPY marker while the male samples will show a single peak which would symbolize the presence of any of the markers of the TSPY gene family. The high copy number would prove to be a merit in sex determination as it increases the sensitivity of this marker. Based on results of a study done for determination of male DNA after intense kissing, the sensitivity of TSPY marker has been proved for sex determination [58]. A similar study on ≤ 3 years old filter paper stored samples concluded that, TSPY is a better marker than SRY as 50% of the samples yielded a positive result with TSPY in comparison to 30% results obtained with SRY [20]. Further studies reinforced the fact that higher copy number in TSPY makes it sensitive and yield better results in forensic samples as compared to SRY which has only one copy. Such studies also established the high sensitivity of TSPY [11].
Giachini et al. [39] reported considerable variation in TSPY copy number in different haplogroups of Italian population. A study on sensitivity of TSPY marker confirmed that, TSPY could give results in DNA as low as 0.004 ng (about 1 sperm cell) whereas SRY could work only up to 0.08 ng (about 25 sperm cells) [53]. Additionally, the efficiency of TSPY in analyzing samples with female DNA as high as 500 times was also found remarkable. The amplification of TSPY was also efficient in the post-coitus samples. It could be amplified till 72 h while SRY could give results only up to 24–48 h. Reports have also shown the detection of TSPY 7 days post-coitus [53] showing superiority of this marker over others for sex determination.
DXYS156
DXY156 is a Penta nucleotide [(TAAAA)n] repeat multi allelic marker located in a long interspersed element (LINE) on both X and Y chromosome, which was first isolated and mapped by Chen et al. [22]. Linkage analysis can be used to map this Penta nucleotide marker to the long arm of X chromosome. Additionally, a sequence homologue is also present at the short arm of Y chromosome [67]. The X and Y counterparts of this marker vary in length i.e. no. of repeat motifs. It has been reported that, 11 repeats or higher are found in locus at Y amplicons while X locus consist of 10 or less number of repeats [115]. In this regard, Cali et al. [19] reported an Adenine insertion in the fourth repeat at the Y locus which could differentiate the X and Y counterparts effectively. They also recommended involving the DXYS156 marker in multiplex analysis kits, for forensic and archeological causes, as this marker is present on both X and Y, it would provide an internal positive control and would result in two peaks similar to amelogenin. Due to its multiallelic nature, it shows variations among different populations or different geographic regions. Reports also suggest the utility of this marker for ethnic studies as well as to study specific patterns of the same allele [63].
Additionally, DXYS156 is highly polymorphic for Indian population. Different population groups exhibit different degrees of variability at Y locus for this marker, whereas, X locus has limited variability as compared to Y locus [88]. This marker does not undergo recombination and hence any mutation in the Y locus tends to accumulate over time. This contributes to the generation of genetic diversity among various ethnic groups of a population [19].
SNPs
Single nucleotide polymorphisms (SNPs) are the most abundantly found variations in the human genome which are present both in coding as well as non-coding regions and account for almost 85% of variations in humans. About 1.9 million SNPs have been registered in the databases till date, proving the relative abundance as well as promising applications of these markers [31]. The added advantages provided by SNP analysis are the low fragment size of 60–80 bp, cost effectiveness, the speed of analysis and huge range of multiplexing [35]. Though SNPs are not as discriminatory as the STR markers, but they promise better results in cases of mass disasters or missing person analysis where reference samples may not be available [131]. Another advantage of SNPs assay is their usefulness in lineage studies using markers on mtDNA or Y chromosome as well as the analysis of phenotypic information based on the SNPs results [69, 75].
SNP markers have been successfully used for human identification as well as gender determination. The advantage of using SNPs for sex determination includes the requirement of lesser sized DNA fragment which is useful in case of degraded forensic samples [68]. In this regard, Wei et al. [121] constructed a SNP panel for ABO blood grouping as well as sex determination. Allwood and Harbison [2] developed a method of sex determination which used single base extension primers to flag Y chromosome regions. This method was found robust on all different aspects of forensic importance like sensitivity and mixed analysis. Recently, a mini-multiplex SNaPshot assay was developed for analysis of degraded samples which included five SNPs for sex determination present on Y chromosome [8]. The superiority of using Y-SNPs over Y STRs include their low mutation rate which can be as low as 100,000 times thus providing a concrete ancestry signature [62, 87]. Additionally, 25 SNPs markers have been analyzed on X chromosome for sexing as well as genetic homogeneity study of Mediterranean populations [114]. In this regard, a novel method has been discovered by Masuyama et al. [82] for accurate sex determination from as low as 20 pg DNA samples by targeting SNPs in the amelogenin gene and sex determining regions on Y chromosome. Another method targeting known X and Y SNP markers called as TriXY-Homogeneous genetic sexing is also being used for sex determination in degraded samples as well as in prenatal diagnosis of gender related birth defects [79]. A recent discovery has suggested the use of SNPs in the genes GATA4 and WWOX for sex determination as well as progression of cancer [70].
However, in current scenario SNPs are categorically being used for forensic phenotyping as they can be sued for the prediction of ethnicity. This ability of SNPs becomes instrumental in cases where there are no suspects [51]. Having highlighted the importance and efficacy of SNPs it’s important to note that SNPs have several benefits but they can’t substitute STRs altogether. Still their role becomes imperative like mtDNA analysis and Y SNPs as they can give you supportive genetic information [17].
DYZ1/Alu assay and others
DYZ1 is a loci present on the long arm of Y chromosome, which may have up to 4000 copies. The high copy number provides an efficient male specific assay. The loci is absent in female samples, thus generating a sex differentiation among humans. The qRT-PCR assays using DYZ1/Alu is considered to be 1500 times more effective than amelogenin assay [34].
About 1 million copies of Alu are present in DNA consisting 10% of the total genome [84]. Microchip electrophoretic sorting of five polymorphic Alu insertions (RC5, A1, PV92, TPA and ACE) have been reported for human ethnicity determination, whereas, two monomorphic Alu insertions (AluSTXa and AluSTYa) for sex typing [92]. Rahman et al. [98] found DYZ1 efficient in discriminating between tissue samples of same individual and samples from sibling males [98]. In this regard, tissue specific individualization is unique to DYZ1 and has high forensic utility. Moreover, its utility in diagnosis of mosaicism gives it an upper edge over all other markers [98]. Sequence and copy number variation of DYZ1 have been reported in prostate cancer biopsied samples [125].
Additionally, pyrosequencing of short PCR products present on amelogenin have also been regarded as a useful tool for sex determination in degraded samples [117]. A novel technique targeting short intergenic sequences (< 50 bp) on both gonosomes has been developed recently called as triplex for the X and Y chromosome (TriXY) [79].
Next generation sequencing
With the advancements in molecular biology, Next Generation Sequencing (NGS) techniques now provide a unique and conclusive result regarding sex determination of any sample irrespective of the nature, condition and presence/absence of genetic disorders. The massive parallel reactions and advanced computational methods make NGS the technology of future. The technology comes with added advantages of reduced costs and enhanced speed. Additionally, Rockenbauer et al. [100] established that many nucleotide substitutions, deletions and insertions are not detected by currently practiced capillary electrophoresis based STR analysis. However, the same can be detected by NGS as it focuses on both the numbers and polymorphisms of repeats.
Detection of a male individual as well as the Y copy number variation (Y-CNV) across the whole chromosome has been studied in much detail with its forensic relevance [81]. Additionally, many sex markers can be targeted to identify sex of an individual rapidly using SEXCMD to human whole exome or RNA sequence datasets using NGS [56]. Application of NGS has also been proven to be useful for sex determination in cases with abnormalities in sex chromosomes such as premature ovarian failure (POF) and sex reversal cases [74]. In this regard, patients with sexual disorder due to mutation in CBX2 gene were categorically characterized for their correct sex using DamID-NGS approach [9]. A recent study emphasized the combinatorial use of NGS, capillary electrophoresis and pyrosequencing, collectively called as ‘NGS+’ for Y-STRs and Y-SNPs typing for sex determination of biological samples at optimum level [96]. Massively parallel sequencing also provides a way to sequence all the available genetic material and type various different kinds of markers in a single run including the sexing makers [5]. The major advantage of using massively parallel sequencing is its capacity to provide results on sex determination using the highly degraded DNA in which the conventional capillary electrophoresis based processes fail to yield results [118].
Alignment of B-allele frequency of X chromosome with reference allele frequency of X chromosome can rapidly identify the sex of the individual from whole genome sequence data [56]. Due to sequence homology of mammalian X and Y chromosomes, Webster et al. [120] developed a statistical tool to minimize technical artifacts in genomic data for reliable sex determination by NGS. NGS has also been reported to be used for reliable sex determination in cases with atypical chromosomal, gonadal or anatomical sex [50]. Besides sex determination, assessment of copy number variation using NGS data is useful in determining effect of azoospermia factor (AZF) loci on male fertility, determination of TSPY copy number variation and detection of AMELY deletion [81]. Additionally, the currently available NGS kits for forensic application such as ForenSeq DNA Signature Prep Kit (Illumina) and HID-Ion AmpliSeq Identity Panel (Ion Torrent), contain additional sex determining markers besides amelogenin increasing their usefulness [3]. Though inception of new and more relevant SNP markers for sex determination by NGS is still in its infancy [104], requirement of scanty amount of DNA sample as low as 62 pg for this purpose is an added advantage [129].
Future challenges of sex determination: chimerism, legal-biological discrepancies and genetic disorders
The future of forensic genetic typing would largely be based on advanced technologies like next generation sequencing. In this regard, RNA markers for sex determination using SNPs are also going to come into the mainstream forensics very soon. SNPs may dominate the sex determination field due to their rapidity and sensitivity. Sex determination in mass disaster cases and conditions with limited samples, would be made possible by SNP markers and the next generation sequencing technology. Additionally, the next generation sequencing technology is going to overturn the capillary based genotyping for sex determination in the upcoming years with faster, contamination free and automatic results with meager amount of DNA samples. The future challenges have been described below.
Chimerism, the presence of genetically varying population of cells in an individual mostly arises due to bone marrow transplant [65]. In this regard, newly transplanted bone marrow migrates to other long bones and is subsequently grafted there followed by the production of new cells with the same genetic lineage as that of the donor; thus, the DNA profiling of the individual differs depending on the sample taken i.e. blood, hair or saliva [52]. With regard to gender determination, no matter what genetic marker we use, as long as the source is blood, the gender detected would be that of the donor instead of the patient [111]. In this respect, recently occurred allogeneic blood transfusion as well as pregnancy with male child may lead to microchimerism which might adversely affect the molecular sex determination using any advanced technique [12].
In the modern era of gender revolutions, genetic sex and gender may not necessarily be the same. On the basis of genetics an offspring can either be a male, female or intersex in case of genetic disorders. But gender is a social term encompassing the individual choices and societal prejudices. A person born as female might identify herself as a male or vice versa and this complicates the issue of gender based identification [107]. On the birth of an intersex child, the parents knowingly or unknowingly assign a gender to the child, which may create a problem in the long run. There occurs a contradiction between the legal gender and biological gender in such cases of intersex and other related issues [55]. In this regard, the gender identity disorder (GID) may also present issues for societal acceptance of molecular sex determination results [99]. Furthermore, people with GID, identify themselves as the opposite sex and often undergo gender transformation surgery [46]. In cases of altered gender, there will be incongruity between the social gender and the biological gender. In the light of increasing instances of people undergoing such surgeries and openly accepting their self-proclaimed sexual identities, sex determination using genetic basis is going to face challenges in near future. The cases of trisomy are most common as XXY, XXX or XYY in humans [23]. On similar lines, Turner syndrome i.e. XO is also witnessed frequently [64]. In such cases the amelogenin or any other sex determining marker will result in correct analysis of genotypic sex but it may not coincide with the legal gender of the person.
Additionally, cases of disorders of sex development such as Androgen Insensitivity syndrome, Gonadal dysgenesis and 5-alpha (2)-reductase-deficiency (ARD) may result in difference in genotypic and phenotypic sex due to abnormalities in the genitalia development [38]. In this regard, 8 SRY positive female athletes were reported among 3,387 cases studied [29]. It can be understood how such abnormalities can have profound effects in the forensic arena. As the gender of the reference samples need to be matched with the registered gender to conclude the involvement, any discrepancy in the gender determination can have a wide impact on the forensic and legal outcomes.
Conclusion
Amelogenin has been the touchstone for sex determination for more than a decade. However, numerous studies have shown the loopholes concerned with amelogenin based sex determination. The cause of such anomalies can be mutations and/or deletions in the Y chromosome. The percentage of such anomalies is very low considering the population sizes but in forensics science even a low percentage of discrepancies matter. An unattended case of Y deletion can lead to false identification of a male as female and hence lead to exclusion of a suspect. Various markers and some advanced technologies like the pyrosequencing and NGS are promising alternatives to using the amelogenin marker. Additionally, use of SNPs, DYZ1/Alu assay can also provide useful information regarding the molecular sex of an individual. Rejecting amelogenin is not an option, multiplex assays that include at least one different marker for sex determination is the solution for this problem. The best marker would have high differentiating power, better sensitivity, good results in mixture analysis and an internal positive control i.e. an X homologue. SNPs seem like the most promising marker for the future of forensic sex determination considering its ability for multiplex, automation and time benefits as they would be a part of the next generation sequencing technologies. In the era of gender revolutions, the marker incorporated to aid amelogenin in sex determination should also propose a solution for determining the gender of individual with incongruent biological and legal sex. With genetic revolutions in mind, the contrasting chromosomal sex and gender complicates the situation and hence, molecular basis of sex determination plays a pivotal role in solving these discrepancies with an added advantage. Moreover, further studies of haplotype groups having higher Y deletion frequencies such as J2, are needed to understand this anomaly in a better way.
References
Álvarez-Sandoval BA, Manzanilla LR, Montiel R (2014) Sex determination in highly fragmented human DNA by high-resolution melting (HRM) analysis. PLoS ONE 9:e104629
Allwood JS, Harbison SA (2015) “YFlag” a single-base extension primer based method for gender determination. J Forensic Sci 60:142–146
Alvarez-Cubero MJ, Saiz M, Martínez-García B, Sayalero SM, Entrala C, Lorente JA, Martinez-Gonzalez LJ (2017) Next generation sequencing: an application in forensic sciences? Ann Hum Biol 44:581–592
Alves C, Coelho M, Rocha J, Amorim A (2006) The Amelogenin locus displays a high frequency of X homologue failures in São Tomé Island (West Africa). Int Cong Ser 1288:271–273
Ambers AD, Churchill JD, King JL, Stoljarova M, Gill-King H, Assidi M, Abu-Elmagd M, Buhmeida A, Budowle B (2016) More comprehensive forensic genetic marker analyses for accurate human remains identification using massively parallel DNA sequencing. BMC Genomics 17:750
Arun Kumar GP, Soria-Hernanz DF, Kavitha VJ, Arun VS, Syama A, Ashokan KS, Gandhirajan KT, Vijayakumar K, Narayanan M, Jayalakshmi M, Ziegle JS, Royyuru AK, Parida L, Wells RS, Renfrew C, Schurr TG, Smith CT, Platt DE, Pitchappan T (2012) Population differentiation of southern Indian male lineages correlates with agricultural expansions predating the caste system. The Genographic Consortium. PLOS ONE 8:7
Bansal AK, Shetty DC, Bindal R, Pathak A (2012) Amelogenin: a novel protein with diverse applications in genetic and molecular profiling. J Oral Maxillofac Pathol 16:395–399
Bardan F, Higgins D, Austin JJ (2018) A mini-multiplex SNaPshot assay for the triage of degraded human DNA. Forensic Sci Int Genet 34:62–70
Bashamboo A, McElreavey K (2016) Mechanism of sex determination in humans: insights from disorders of sex development. Sex Dev 10:313–325
Bernard P, Sim H, Knower K, Vilain E, Harley V (2008) Human SRY inhibits β-catenin-mediated transcription. Int J Biochem Cell Biol 40:2889–2990
Blagodatskikh EG, Nikitin AG, Seregin YA, Blagodatskikh KA, Nosikov VV (2010) Sex determination in biological specimens using the DYS14 marker. Mol Biol 44:568–570
Bloch EM, Jackman RP, Lee TH, Busch MP (2013) Transfusion associated microchimerism: the hybrid within. Transf Med Rev 27:10–20
Borovko S, Shyla A, Korban V, Borovko A (2015) Amelogenin test abnormalities revealed in Belarusian population during forensic DNA analysis. Forensic Sci Int Genet 15:98–104
Bruzek J, Murail P (2006) Methodology and reliability of sex determination from the skeleton. In: Schmitt A, Cunha E, Pinheiro J (eds) Forensic anthropol med. Humana Press, Totowa
Butler E, Li R (2014) Genetic markers for sex identification in forensic DNA analysis. J Forensic Invest 2:1–10
Butler JM (2015) The future of forensic DNA analysis. Philos Trans R Soc Lond B 370:20140252
Butler JM, Coble MD, Vallone PM (2007) STRs vs. SNPs: thoughts on the future of forensic DNA testing. Forensic Sci Med Pathol 3:200–205
Cadenas AM, Regueiro M, Gayden T, Singh N, Zhivotovsky LA, Underhill PA, Herrera RJ (2007) Male amelogenin dropouts: phylogenetic context, origins and implications. Forensic Sci Int 166:155–163
Calì F, Forster P, Kersting C, Mirisola MG, Anna RD, Leo GD (2002) DXYS156: a multi-purpose short tandem repeat locus for determination of sex, paternal and maternal geographic origins and DNA fingerprinting. Int J Leg Med 116:133–138
Campos EA, Pitta DR, Costa FA, Campos VM, Yela D, Fernandes A (2014) DNA extraction from filter-paper spots of vaginal samples collected after sexual violence. Int J Gynecol Obs 126:23–27
Chang YM, Burgoyne LA, Both K (2003) Higher failures of amelogenin sex test in an Indian population group. J Forensic Sci 48:1309–1313
Chen H, Lowther W, Avramopoulos D, Antonarakis SE (1994) Homologous loci DXYS156X and DXYS156Y contain a polymorphic pentanucleotide repeat (TAAAA)n and map to human X and Y chromosomes. Hum Mutat 4:208–211
Chen X, Williams-Burris SM, McClusky R, Ngun TC, Ghahramani N, Barseghyan H, Reue K, Vilain E, Arnold AP (2013) The sex chromosome trisomy mouse model of XXY and XYY: metabolism and motor performance. Biol Sex Diff 4:15
Codina AE, Niederstätter H, Parson W (2009) GenderPlex a PCR multiplex for reliable gender determination of degraded human DNA samples and complex gender constellations. Int J Leg Med 123:459–464
Crawford PJM, Aldred M, Zupan AB (2007) Amelogenesis imperfect. Orphanet J Rare Dis 2:17
Dash HR, Das S (2018) Microbial degradation of forensic samples of biological origin: potential threat to human DNA typing. Mol Biotechnol 60:141–153
Deagle BE, Eveson JP, Jarman SN (2006) Quantification of damage in DNA recovered from highly degraded samples- a case study on DNA in faeces. Front Zool 3:11
Dechend F, Williams G, Skawran B, Schubert S, Krawczak M, Tyler-Smith C, Schmidtke J (2000) TSPY variants in six loci on the human Y chromosome. Cytogenet Cell Genet 91:67–71
Dickinson BD, Genel M, Robinowitz CB, Turner PL, Woods GL (2002) Gender verification of female olympic athletes. Med Sci Sports Exerc 34:1539–1542
Dixit S, Shrivastava P, Kumawat RK, Kaitholia K, Dash HR, Sharma H, Choubey G (2019) Forensic genetic analysis of population of Madhya Pradesh with PowerPlex Fusion 6C™ Multiplex System. Int J Leg Med 133:803–805
Doddamani D, Khan AW, Katta MAVSK, Agarwal G, Thudi M, Ruperao P, Edwards D, Varshney RK (2015) CicArVarDB: SNP and InDel database for advancing genetics research and breeding applications in chickpea. Database (Oxford) 2015:bav078
Drobnič K (2006) A new primer set in a SRY gene for sex identification. Int Cong Ser 1288:268–270
Dutta P, Bhosale S, Singh R, Gubrellay P, Patil J, Sehdev B, Bhagat S, Bansal T (2017) Amelogenin gene:the pioneer in gender determination from forensic dental samples. J Clin Diagn Res 11:ZC56–ZC59
Fazi A, Gobeski B, Foran D (2014) Development of two highly sensitive forensic sex determination assays based on human DYZ1 and Alu repetitive DNA elements. Electrophoresis 35:3028–3035
Fernández ME, Goszczynski DE, Lirón JP, Villegas-Castagnasso EE, Carino MH, Ripoli MV, Rogberg-Muñoz A, Posik DM, Peral-García P, Giovambattista G (2013) Comparison of the effectiveness of microsatellites and SNP panels for genetic identification, traceability and assessment of parentage in an inbred Angus herd. Genet Mol Biol 36:185–191
Fincham AG, Moradian-Oldak J, Simmer JP (1999) The structural biology of the developing dental enamel matrix. J Struct Biol 126:270–299
Fontanesi L, Scotti E, Russo V (2008) Differences of the porcine amelogenin X and Y chromosome genes (AMELX and AMELY) and their application for sex determination in pigs. Mol Reprod Dev 75:1662–1668
Gangaher A, Chauhan V, Jyotsna VP, Mehta M (2016) Gender identity and gender of rearing in 46 XY disorders of sexual development. Ind J Endocrinol Metabol 20:536–541
Giachini C, Nuti F, Turner DJ, Laface I, Xue Y, Daguin F, Forti G, Tyler-Smith C, Krausz C (2015) TSPY1 copy number variation influences spermatogenesis and shows differences among Y lineages. J Clin Endocrinol Metabol 94:4016–4022
Gibbon V, Paximadis M, Strkalj G, Ruff P, Penny C (2009) Novel methods of molecular sex identification from skeletal tissue using the amelogenin gene. Forensic Sci Int Genet 3:74–79
Gilder JR, Inman K, Shields W, Krane DE (2010) Magnitude-dependent variation in peak height balance at heterozygous STR loci. Int J Leg Med 125:87–94
Gnanavel S, Hussain S (2017) X-linked icthyosis and neurodevelopmental disorders: a case report and review of literature. Asian J Psychol 28:1–2
Graves JAM (2002) The rise and fall of SRY. Trends Genet 18:259–264
Hansson O, Egeland T, Gill P (2017) Characterization of degradation and heterozygote balance by simulation of the forensic DNA analysis process. Int J Leg Med 131:303–317
Hares DR (2015) Selection and implementation of expanded CODIS core loci in the United States. Forensic Sci Int Genet 17:33–34
Hess J, Neto RR, Panic L, Rübben H, Senf W (2014) Satisfaction with male-to-female gender reassignment surgery: results of a retrospective analysis. Deutsches Ärzteblatt Int 111:795–801
Hobson GM, Gibson CW, Aragon M, Yuan Z, Davis-Williams A, Banser L, Kirkham J, Brook AH (2009) A large X-chromosomal deletion is associated with microphthalmia with linear skin defects (MLS) and amelogenesisimperfecta (XAI). Am J Med Genet A 149:1698–1705
Hrovatin K, Kunej T (2018) Genetic sex determination assays in 53 mammalian species: literature analysis and guidelines for reporting standardization. Ecol Evol 8:1009–1018
Hu JCC, Chun YHP, Al Hazzazzi T, Simmer JP (2007) Enamel formation and amelogenesis imperfecta. Cells Tissue Org 186:78–85
Hughes LA, McKay-Bounford K, Webb EA, Dasani P, Clokie S, Chandran H, McCarthy L, Mohamed Z, Kirk JMW, Krone NP, Allen S, Cole TRP (2019) Next generation sequencing (NGS) to improve the diagnosis and management of patients with disorders of sex development (DSD). Endocr Connect 8:100–110
Irwin J, Just R, Scheible M, Loreille O (2011) Assessing the potential of next generation sequencing technologies for missing persons identification efforts. Forensic Sci Int Genet Suppl Ser 3:e447–e448
Jacewicz R, Lewandowski K, Matysek JR, Jedrzejczyk M, Komarnicki M, Berent J (2013) Genetic investigation of biological materials from patients after stem cell transplantation based on autosomal as well as Y-chromosomal markers. Int J Leg Med 127:359–362
Jacot TA, Zalenskaya I, Mauck C, Archer DF, Doncel GF (2013) TSPY4 is a novel sperm-specific biomarker of semen exposure in human cervicovaginal fluids; Potential use in HIV prevention and contraception studies. Contraception 88:387–395
Jakob S, Lovell-Badge R (2011) Sex determination and the control of Sox9 expression in mammals. FEBS J 278:1002–1009
Jenkins TM, Short SE (2017) Negotiating intersex: a case for revising the theory of social diagnosis. Soc Sci Med 175:91–98
Jeong S, Kim J, Park W, Jeon H, Kim N (2017) SEXCMD: development and validation of sex marker sequences for whole-exome/genome and RNA sequencing. PLoS ONE 12:e0184087
Jobling MA, Lo ICC, Turner DJ, Bowden GR, Lee AC, Xue Y, Carvalho-silva D, Hurles ME, Adams SM, Meng CY, Guanti G, Mckeown B, Oorschot RAHV, Mitchell RJ, Knijff PD, Tyler-smith C, Parkin EJ (2007) Structural variation on the short arm of the human Y chromosome: recurrent multigene deletions encompassing Amelogenin Y. Hum Mol Genet 16:307–316
Kamodyová N, Durdiaková J, Celec P, Sedláčková T, Repiská G, Sviežená B, Minárik G (2013) Prevalence and persistence of male DNA identified in mixed saliva samples after intense kissing. Forensic Sci Int Genet 7:124–128
Kareem MA, Hussein AO, Hameed IH (2015) Y-Chromosome short tandem repeat, typing technology, locus information and allele frequency in different population: a review. Afr J Biotechnol 14:2175–2178
Kashyap VK, Sahoo S, Sitalaximi T, Trivedi R (2006) Deletions in the Y-derived amelogenin gene fragment in the Indian population. BMC Med Genet 7:37
Kastelic V, Budowle B, Drobnič K (2009) Validation of SRY marker for forensic casework analysis. J Forensic Sci 54:551–555
Kayser M (2017) Forensic use of Y-chromosome DNA: a general overview. Hum Genet 136:621–635
Kersting C, Hohoff C, Rolf B, Brinkmann B (2001) Pentanucleotide short tandem repeat locus DXYS156 displays different patterns of variations in human populations. Croat Med J 42:310–314
Kesler SR (2007) Turner Syndrome. Child Adolesc Psychiatr Clin N Am 16:709–722
Khan F, Agarwal A, Agrawal S (2004) Significance of chimerism in hematopoietic stem cell transplantation: new variations on an old theme. Bone Marrow Transpl 34:1–12
Kida M, Sakiyama Y, Matsuda A, Takabayashi S, Ochi H, Sekiguchi H, Minamitake S, Ariga T (2007) A novel missense mutation (p. P52R) in amelogenin gene causing X-linked amelogenesisimperfecta. J Dent Res 86:69–72
Kim HS, Crow TJ (1998) Human DXYS156 of pentanucleotide repeat (TAAAA)n: chromosomal localization by somatic hybrid mapping and sequencing analysis. Cytogenet Cell Genet 83:54–55
Kim JJ, Han BG, Lee HI, Yoo HW, Lee JK (2010) Development of SNP-based human identification system. Int J Leg Med 124:125–131
Kivisild T (2017) The study of human Y chromosome variation through ancient DNA. Hum Genet 136:529–546
Kristiansen W, Karlsson R, Rounge TB, Whitington T, Andreassen BK, Magnusson PK, Fosså SD, Adami HO, Turnbull C, Haugen TB, Grotmol T, Wiklund F (2015) Two new loci and gene sets related to sex determination and cancer progression are associated with susceptibility to testicular germ cell tumor. Hum Mol Genet 24:4138–4146
Kumagai R, Sasaki Y, Tokuta T, Biwasaka H, Aoki Y (2008) Case report DNA analysis of family members with deletion in Yp11, 2 region containing amelogeninlocus. Leg Med (Tokyo) 10:39–42
Lattanzi W, Di Giacomo MC, Lenato GM, Chimienti G, Voglino G, Resta N, Pepe G, Guanti G (2005) A large interstitial deletion encompassing the amelogenin gene on the short arm of the Y chromosome. Hum Genet 116:395–401
Laverde L (2013) Sex determination problems in forensic genetic analysis. Forensic Sci Int Genet Suppl Ser 4:e350–e351
Lee Y, Kim C, Joon Y, Pyun JA, Kwack KB (2016) Next generation sequencing identifies abnormal Y chromosome and candidate causal variants in premature ovarian failure patients. Genome 108:209–215
Lorenz AJ, Hamblin MT (2010) Performance of single nucleotide polymorphisms versus haplotypes for genome-wide association analysis in Barley. PLoS ONE 5:e14079
Luce C, Montpetit S, Gangitano D, O’Donnell P (2009) Validation of the AMPFlSTR MiniFiler PCR amplification kit for use in forensic casework. J Forensic Sci 54:1046–1054
Ma Y, Kuang JZ, Zhang J, Wang GM, Wang YJ, Jin WM, Hou YP (2012) Y chromosome interstitial deletion induced Y-STR allele dropout in AMELY-negative individuals. Int J Leg Med 126:713–724
Maciejewska A, Pawłowski R (2009) A rare mutation in the primer binding region of the Amelogenin X homologue gene. Forensic Sci Int Genet 3:265–267
Madel MB, Niederstätter H, Parson W (2016) TriXY-Homogeneous genetic sexing of highly degraded forensic samples including hair shafts. Forensic Sci Int Genet 25:166–174
Makiyan Z (2016) Studies of gonadal sex differentiation. Organogenesis 12:42–51
Massaia A, Xue Y (2017) Human Y chromosome copy number variation in the next generation sequencing era and beyond. Hum Genet 136:591–603
Masuyama K, Shojo H, Nakanishi H, Inokuchi S, Adachi N (2017) Sex determination from fragmented and degenerated DNA by amplified product length polymorphism bidirectional SNP analysis of Amelogenin and SRY genes. PLoS ONE 12:e0169348
Michael A, Brauner P (2004) Erroneous gender identification by the amelogenin sex test. J Forensic Sci 49:258–259
Mighell A, Markham A, Robinson P (1997) Alu sequences. FEBS Lett 417:1–5
Mitchell RJ, Kreskas M, Baxter E, Buffalino L, Van Oorschot RAH (2006) An investigation of sequence deletions of amelogenin (AMELY), a Y-chromosome locus commonly used for gender determination. Ann Hum Biol 33:227–240
Morikawa T, Yamamoto Y, Miyaishi S (2011) A new method for sex determination based on detection of sex determining region Y, steroid sulfatase and amelogenin gene regions with simultaneous amplification of their homologous sequences by a multiplex polymerase chain reaction. Acta Med Okayama 65:113–122
Morimoto C, Manabe S, Fujimoto S, Hamano Y, Tamaki K (2018) Discrimination of relationships with the same degree of kinship using chromosomal sharing patterns estimated from high-density SNPs. Forensic Sci Int Genet 33:10–16
Mukerjee S, Mukherjee M, Ghosh T, Kalpana D, Sharma AK (2013) Differential pattern of genetic variability at the DXYS156 locus on homologous regions of X and y chromosomes in Indian population and its forensic implications. Int J Leg Med 127:1–6
Murphy E (2018) Forensic DNA typing. Ann Rev Criminol 1:497–515
Muthusamy D (2017) Determination of sex of an individual by identifying SRY gene through PCR analysis of DNA extracted from incinerated teeth- a single blind pilot study. Int J Forensic Sci 2:000128
Nakahori Y, Takenaka O, Nakagome Y (1991) A human X-Y homologous region encodes “amelogenin”. Genomics 9:264–269
Njoroge SK, Witek MA, Hupert ML, Soper SA (2010) Microchip electrophoresis of Alu elements for gender determination and inference of human ethnic origin. Electrophoresis 31:981–990
Ou X, Chen W, Chen H, Zhao F, Zheng J, Tong D, Chen Y, Chen A, Sun H (2012) Null alleles of the X and Y chromosomal amelogenin gene in a Chinese population. Int J Leg Med 126:513–518
Oyama N, Satoh M, Iwatsuki K, Kaneko F (2000) Novel point mutations in the steroid sulfatase gene in patients with X-linked Ichthyosis: transfection analysis using the mutated genes. J Invest Dermatol 114:1195–1199
Pierce KE, Rice JE, Sanchez JA, Brenner C, Wangh LJ (2000) Real time PCR using molecular beacons for accurate detection of the Y chromosome in single human blastomeres. Mol Hum Reprod 6:1155–1164
Qian X, Hou J, Wang Z, Ye Y, Lang M, Gao T, Liu J, Hou Y (2017) Next generation sequencing plus (NGS+) with Y-chromosomal markers for forensic pedigree searches. Sci Rep 7:11324
Quintana-Murci L, Fellous M (2001) The human Y chromosome: the biological role of a “functional wasteland”. J Biomed Biotechnol 1:18–24
Rahman M, Bashamboo A, Prasad A, Pathak D, Ali S (2004) Organizational variation of DYZ1 repeat sequences on the human Y chromosome and its diagnostic potentials. DNA Cell Biol 23:561–571
Rathi A, Bhatia MS (2014) Management challenges in a case of gender identity disorder. Ind Psychol J 23:157–159
Rockenbauer E, Hansen S, Mikkelsen M, Børsting C, Morling N (2014) Characterization of mutations and sequence variants in the D21S11 locus by next generation sequencing. Forensic Sci Int Genet 8:68–72
Roffey PE, Eckhoff CI, Kuhl JL (2000) A rare mutation in the amelogenin gene and its potential investigative ramifications. J Forensic Sci 45:1016–1019
Santos FR, Pandya A, Tyler-Smith C (1998) Reliability of DNA-based sex tests. Nat Genet 18:103–103
Sasaki S, Shimokawa H (1995) The amelognin gene. Int J Dev Biol 39:127–133
Seo S, King J, Warshauer D, Davis C, Ge J, Budowle B (2013) Single nucleotide polymorphism typing with massively parallel sequencing for human identification. Int J Leg Med 127:1079–1086
Settin A, Elsobky E, Hammad A, Al-Erany A (2008) Rapid sex determination using PCR technique compared to classic cytogenetics. Int J Health Sci 2:49–52
Shadrach B, Commane M, Hren C, Warshawsky I (2004) A rare mutation in the primer binding region of the amelogenin gene can interfere with gender identification. J Mol Diagn 6:401–405
Short SE, Yang YC, Jenkins TM (2013) Sex, gender, genetics, and health. Am J Public Health Res 103:S93–S101
Smith CEL, Murillo G, Brookes SJ, Poulter JA, Silva S, Kirkham J, Inglehearn CF, Mighell AJ (2016) Deletion of amelotin exons 3-6 is associated with amelogenesis imperfecta. Hum Mol Genet 25:3578–3587
Steinlechner M, Berger B, Niederstätter H, Parson W (2002) Rare failures in the amelogenin sex test. Int J Leg Med 116:117–120
Sullivan KM, Mannucci A, Kimpton CP, Gill P (1993) A rapid and quantitative DNA sex test: fluorescence-based PCR analysis of X-Y homologous gene amelogenin. Biotechniques 15:636–638
Swierczynski SL, Hafez MJ, Philips J, Higman MA, Berg KD, Murphy KM (2005) Bone marrow engraftment analysis after granulocyte transfusion. J Mol Diagn 7:422–426
Takayama T, Takada N, Suzuki R, Nagaoka S, Watanabe Y, Kumagai R, Aoki Y, Butler JM (2009) Determination of deleted regions from Yp11.2 of an amelogenin negative male. Leg Med 11:S578–S580
Thangaraj K, Reddy AG, Singh L (2002) Is the amelogenin gene reliable for gender identification in forensic casework and prenatal diagnosis? Int J Leg Med 116:121–123
Tomas C, Sanchez JJ, Barbaro A, Brandt-Casadevall C, Hernandez A, Dhiab MB, Ramon M, Morling N (2008) X-chromosome SNP analyses in 11 human Mediterranean populations show a high overall genetic homogeneity except in North-west Africans (Moroccans). BMC Evol Biol 8:75
Torres-Rodríguez M, Martínez-Cortes G, Páez-Riberos LA, Sandoval L, Muñoz-Valle JF, Ceballos-Quintal JM, Pinto-Escalante D, Rangel-Villalobos H (2006) Forensic potential of the STR DXYS156 in Mexican populations: inference of X-linked allele null. Leg Med 8:52–54
Trent S, Davies W (2013) Cognitive, behavioural and psychiatric phenotypes associated with steroid sulfatase deficiency. World J Transl Med 2:1–12
Tschentscher F, Frey UH, Bajanowski T (2008) Amelogenin sex determination by pyrosequencing of short PCR products. Int J Leg Med 122:333–335
Tucker T, Marra M, Friedman JM (2009) Massively parallel sequencing: the next big thing in genetic medicine. Am J Hum Genet 85:142–154
Vodicka R, Vrtel R, Dusek L, Singh AR, Krizova K, Svacinova V, Horinova V, Dostal J, Oborna I, Brezinova J, Sobek A, Santavy J (2007) TSPY gene copy number as a potential new risk factor for male infertility. Reprod BioMed Online 14:579–587
Webster TH, Couse M, Grande BM, Karlins E, Phung TN, Richmond PA, Whitford W, Wilson MA (2019) Identifying, understanding, and correcting technical artifacts on the sex chromosomes in next-generation sequencing data. Gigascience. https://doi.org/10.1093/gigascience/giz074
Wei YL, Li CX, Jia J, Hu L, Liu Y (2012) Forensic identification using a multiplex assay of 47 SNPs. J Forensic Sci 57:1448–1456
Westen AA, Kraaijenbrink T, de Medina EAR, Harteveld J, Willemse P, Zuniga SB, van der Gaag KJ, Weiler NEC, Warnaar J, Kayser M, Sijen T, Knijff P (2014) Comparing six commercial autosomal STR kits in a large Dutch population sample. Forensic Sci Int Genet 10:55–63
Xie J, Shao C, Xu H, Zhu W, Liu Z, Tang Q, Zhou Y (2014) Deletion mapping of the regions with AMELY from two Chinese males. Leg Med 16:290–292
Xue Y, Tyler-Smith C (2011) An exceptional gene: evolution of the TSPY gene family in humans and other great apes. Genes (Basel) 2:26–47
Yadav SK, Kumari A, Ali S (2013) Fate of the human Y chromosome linked genes and loci in prostate cancer cell lines DU145 and LNCaP. BMC Genomics 14:323. https://doi.org/10.1186/1471-2164-14-323
Yen PH, Marsh B, Allen E, Tsai SP, Ellison J, Connolly L, Neiswanger K, Shapiro LJ (1988) The human X-linked steroid sulfatase gene and a Y-encoded pseudogene: evidence for an inversion of the Y chromosome during primate evolution. Cell 55:SS1123–SS1135
Yong RYY, Gan LSH, Chang YM, Yap EPH (2007) Molecular characterization of a polymorphic 3-Mb deletion at chromosome Yp11.2 containing the AMELY locus in Singapore and Malaysia populations. Hum Genet 122:237–249
Zehethofer K, Rolf B (2011) A molecular analysis of three amelogenin negative males in two routine paternity tests. Forensic Sci Int Genet 5:550–551
Zeng X, King J, Stoljarova M, Warshauer D, LaRue B, Sajantila A, Patel J, Storts D, Budowle B (2015) High sensitivity multiplex short tandem repeat loci analyses with massively parallel sequencing. Forensic Sci Int Genet 16:38–47
Zhao Z, Khan F, Borkar M, Herrera R, Agrawal S (2009) Presence of three different paternal lineages among North Indians: a study of 560 Y chromosomes. Ann Hum Biol 36:46–59
Ziętkiewicz E, Witt M, Daca P, Żebracka-Gala J, Goniewicz M, Jarząb B, Witt M (2012) Current genetic methodologies in the identification of disaster victims and in forensic analysis. J Appl Genet 53:41–60
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they do not have any conflict of interest.
Ethical approval
This review article does not report any experiments conducted with human participants or with other animals.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Dash, H.R., Rawat, N. & Das, S. Alternatives to amelogenin markers for sex determination in humans and their forensic relevance. Mol Biol Rep 47, 2347–2360 (2020). https://doi.org/10.1007/s11033-020-05268-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05268-y