Abstract
Colorectal cancer (CRC) is among the leading causes of cancer-related mortality worldwide. Compelling evidence suggests that long non-coding RNA (lncRNAs) can control carcinogenesis by regulating various aspects of cell biology. However, limited number of CRC-related lncRNAs has been well characterized. This study was undertaken to investigate the expression pattern of the novel lncRNA-CCHE1 in CRC patients and to examine its correlation with clinicopathological features, ERK/COX-2 pathway and some cell proliferation markers in order to gain biological insights on its role in CRC pathogenesis. Colon cancer specimens with their adjacent non-cancerous tissues were taken from 60 patients with primary CRC. LncRNA-CCHE1 relative expression was assessed using quantitative real-time RT-PCR. P-ERK ½ and cyclin D1 levels were estimated by ELISA. COX-2 and proliferating cell nuclear antigen (PCNA) expression were assessed immunohistochemically. lncRNA-CCHE1 expression was upregulated in CRC tissues compared to adjacent non-cancerous tissues, and was significantly associated with larger tumor size, less differentiated histology, advanced dukes’ stage, positive lymph node involvement and vascular invasion. It also showed a significant positive correlation with the expression of p-ERK1/2, COX-2 as well as cyclin D1and PCNA (as markers for cell proliferation). These findings signify that lncRNA-CCHE1 is a key oncogene possibly involved in CRC development and progression by modulating ERK/COX-2 pathway and cell proliferation activity. Our study also provides a rationale for potential use of lncRNA-CCHE1 as a novel prognostic marker, and opens the door for the development of lncRNA-CCHE1-directed therapeutic approaches for CRC patients.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is among the leading causes of cancer-related mortality worldwide [1]. CRC development and progression involve complex multi-factorial steps, in which aberrant gene expression and consequent deregulated biological pathways play a significant role [2].
Long non-coding RNAs (lncRNAs) are untranslated RNA polymerase II transcripts of > 200 nucleotides that lack an open reading frame [3]. Despite the fact that, according to the human genome sequencing databases, half of the human genome is lncRNA; the functions of the majority of lncRNAs are not fully clear [4].
Nevertheless, recent research in transcriptome profiles has highlighted the role of some lncRNAs as main regulators of gene expression; probably through epigenetic modifications, their interactions with miRNA and proteins, as well as their actions as miRNA precursors or pseudogenes [5]. For instance, previous reports have identified a number of CRC—associated lncRNAs; including HOTAIR, MALAT1, BANCR, H19, and HULC [6]. These lncRNAs could control colorectal carcinogenesis by regulating various aspects of cell biology such as cell proliferation, apoptosis, and invasion [7]. However, the lncRNAs landscape and their overall pathophysiological contributions to CRC remain to be fully elucidated.
Cervical carcinoma high-expressed lncRNA 1 (lncRNA-CCHE1) is a newly discovered lncRNA with 2504 nucleotides in length and located in an intergenic region on chromosome 10. Recently, it was reported to be highly expressed in cervical cancer tissues and was therefore nominated as a molecular biomarker for cervical cancer prognosis [8]. However, the clinical value and biological significance of CCHE1 in colorectal carcinogenesis require further exploration.
Cyclooxygenase-2 (COX-2) is an inducible pro-inflammatory enzyme whose expression is up-regulated at sites of inflammation and in several epithelial cancers, where it is engaged in cell cycle alterations, apoptosis suppression, angiogenesis, and tumor cell invasiveness [9]. COX-2 expression is controlled by multiple signal transduction pathways, including the mitogen-activated protein kinase (MAPK)/ERK pathway [10] which is a significant cell signaling pathway and through its activation, numerous key growth factors and proto-oncogenes can transmit signals promoting tumor cell proliferation and differentiation [11]. Among these proto-oncogenes, cyclin D1 is a key cell-cycle regulator, which controls the G1–S phase progression in mitotic mammalian cells and its dysregulated expression has been linked to cancer development and progression [12].
Since lncRNAs are extensively modulated by multiple transcriptional regulators [13], it is of critical importance to investigate the association between these regulatory mechanisms, such as ERK/COX-2, and their subsequent effects on lncRNA expression and function in various types of cancer.
Therefore, this study was undertaken to investigate the expression pattern of the novel lncRNA-CCHE1 in CRC patients; in the context of some cell cycle and proliferation markers, [including cyclin D1and proliferating cell nuclear antigen (PCNA)], as well as to examine its correlation with ERK/COX-2 pathway and clinicopathological features in order to attain a better understanding for its role in CRC pathogenesis; which might open new avenues for developing lncRNA-directed diagnostic and therapeutic approaches.
Patients and methods
Patients
This study included 60 patients presenting with primary colorectal carcinoma (CRC) admitted to the General Surgery Department of Tanta University Hospital, during the period between March 2016 and March 2018. Patients with other confounding pathologies in the colon, autoimmune disease, acute infection and patients with prior radiotherapy, immunosuppressive, chemotherapeutic drugs, obstructed or perforated CRC, were excluded from the study. All subjects gave their written informed consent before participation. The study protocol was approved by the local ethics committee at Faculty of medicine; Tanta University, and was in accordance with the principles of the Declaration of Helsinki II.
All patients were diagnosed as primary cancer colon by history, clinical examination and investigations including pelvi-abdominal ultrasonography, computed tomography, colonoscopy and tissue biopsy for histopathological study. All patients were prepared for surgery by routine pre-operative investigations (complete blood picture, coagulation profile, blood sugar, as well as liver and kidney functions) and colonic preparation. Next, laparotomy was performed and tumor was evaluated intraoperatively as regards tumor site, size, mobility, lymphatic and vascular spread, then it was surgically removed.
Methods
Colonic tissue specimens
Tumor samples were taken from the invasive edge of freshly resected tumour specimens. Corresponding samples of normal colonic mucosa, enrolled in this study as a control, were taken from the furthest resection margin. Samples were washed with iced cold saline, snap frozen then divided into two parts: one part was immediately stored in at − 80 °C for RNA extraction and immunoassay, while the other part was fixed in 10% formalin for histopathological and immunohistochemical examination.
Colonic tissue homogenization
Tissue samples were homogenized in 20 mM Tris–HCl (pH 7.5), 1 mM DTT, 1 mM MgCl 2 and 1 mM EDTA, and centrifuged at 10,000×g for 10 min at 4 °C.
Preparation of nuclear extract
Nuclear extracts from colonic tissue were prepared using the Nuclear/Cytosol Fractionation Kit (Cat #K266-25, BioVision, Inc., CA, USA) according to the protocol of the manufacturer.
Biochemical analysis
-
(A)
Total proteins Concentrations in the samples were determined according to the method of Bradford with bovine serum albumin as a standard (#Cat no.500-0006, Bio\Rad Protein Assay) [14].
-
(B)
Phosphorylated extracellular signal regulated kinase1/2 (p-ERK1/2) levels in colonic tissue homogenates were assayed using an ELISA kit (Cat # 201-12-6290; SunRed BioTechnology Co., Ltd,. Shan. China) ; as per the manufacturer’s protocols.
-
(C)
Cyclin D1 levels were assayed in nuclear extracts of colonic tissue samples using an ELISA kit (Cat # ab214571; Abcam Biochemicals; Cambridge., UK) ; according to the manufacturer’s instructions.
-
(D)
Quantitative measurement of long non-coding RNA CCHE1 gene expression by quantitative real-time reverse transcription PCR (RT-PCR):
-
i.
RNA extraction Total RNA was extracted from colon cancers and adjacent non-tumor specimens using Gene JET RNA Purification Kit (Thermo Scientific, # K0731, USA) according to the manufacturer’s protocol. The samples were treated with DNase to exclude contaminating genomic DNA. Total RNA concentration and purity were determined by measuring OD260 and OD260/280 ratio, respectively, on a NanoDrop spectrophotometer (NanoDrop Technologies, Inc. Wilmington, USA), RNA was then stored at − 80 °C.
-
ii.
cDNA synthesis was performed using the RevertAid H Minus First Strand cDNA Synthesis kit (Cat#K1632,Thermo Scientific Fermentas, St. Leon-Ro, Germany) according to the manufacturer’s instructions. 10 µl of random hexamer primers (Roche, Mannheim, Germany) were added to 21 µl of RNA which was denatured for 5 min in the thermal cycler (Biometra, USA). The RNA-primer mixture was cooled to 4 °C. The cDNA master mix was prepared according to the kit’s protocol (5 µl of first strand buffer, 10 mM of dNTPs, 1 µl of RNase inhibitor, 1 µl of reverse transcriptase Superscript™ II-RT enzyme and 10 µl of DEPC treated water) and was added to each sample. The total volume of the cDNA master mix for each sample was 19 µl. This was added to 31 µl RNA-primer mixtures resulting in a reaction volume of 50 µl, which was then incubated in the programmed thermal cycler 1 h at 37 °C, followed by inactivation of enzymes at 95 °C for 10 min, and finally cooled at 4 °C. The RNA was reverse transcribed into cDNA which was then stored at − 80 °C.
-
iii.
Real-time quantitative PCR: 1 µl of this cDNA was added to 20 µl reaction mixture of the QuantiTect SYBR-Green PCR kit (Qiagen) and 0.5 µM from the specific primer pairs for of lncRNA-CCHE1. This cDNA was then amplified using StepOnePlus real time PCR system (Applied Biosystem, USA) as follows: Initial denaturation at 95 °C for 10 min was followed by 40 cycles with denaturation at 95 °C for 15 s, annealing at 60 °C for 30 s and extension at 72 °C for 30 s. Primer sequences specific for long non-coding RNA CCHE1 (NCBI GenBank Nucleotide accession # NM_ AK055418.1) were designed according to Zhan et al. [15] as follows: forward 5′- AAGGTCCCAGGATACTCGC-3′ and reverse 5′- GTGTCGTGGACTGGCAAAAT-3′. Primers for GAPDH (NCBI GenBank Nucleotide accession # NM_001357943.1), which was used as a reference to calculate fold change in target gene expression, were: forward 5′-CGCTCTCTGCTCCTCCTGTTC-3′ and reverse 5′-ATCCGTTGACTCCGACCTTCAC-3′. The cycle threshold (Ct) values were calculated for target genes and the reference gene, and relative gene expression was determined using 2−∆∆Ct method [16].
-
i.
Histopathological examination
Histopathological examination was done to determine tumor histological type, grade according to Broders system [17], and tumor stage according to the Dukes system [18].
Immunohistochemical staining
The avidin–biotin complex method was used to detect PCNA and Cox-2. Formalin-fixed and paraffin-embedded tissues were deparaffinized with xylene and rehydrated in ethanol. After quenching of endogenous peroxidase activity with 3% H2O2 for 30 min, the slides were submerged in EDTA antigenic retrieval buffer (pH 8.0) and heated in a microwave oven. Sections were blocked for 30 min at 37 °C with 1% bovine serum albumin (BSA). Rabbit Polyclonal PCNA and Cox-2 antibodies, ready to use, (IHC World, # IW-MA1083 and IW-PA1121), were incubated on slides in a moist chamber overnight at 4 °C. Biotinylated goat anti-mouse secondary antibody was applied for 15 min at 37 °C, followed by incubation with a complex of avidin with horseradish peroxidase for 15 min at 37 °C. Sections were developed with diaminobenzidine and counterstained with hematoxylin. Finally, the slides were sequentially dehydrated in ethanol and mounted.
Scoring immunoreactivity
Stained sections were microscopically examined and scored by a pathologist who was blinded to the clinical data.
-
i.
Scoring of COX-2 expression in tumor cells was done according to the methods of Remmele and Stegner [19]. The intensity of staining was scored as 0 (negative), 1 (weak), 2 (medium), or 3 (strong), and the extent of staining was scored as 0 (0%), 1 (1–25%), 2 (26–50%), 3 (51–75%), and 4 (76–100%), indicating the percentage of positive staining in the carcinoma tissue. Addition of an intensity score (0–3) and an extent score (0–4) resulted in a COX-2 immunoreactivity score (IRS-COX2), which ranged between 0 and 7. For statistical purposes, tumors having a final staining score of ≥ 3 were considered to be positive.
-
ii.
Scoring of PCNA expression in tumor cells was done according to the percentage of positive nuclei [20]. Less than 25% positive was scored as negative, and ≥ 25% was scored as positive.
Statistical analysis
Quantitative data are expressed as mean ± standard deviation (SD).The intergroup variation between two groups was measured using t-test, while variations between three groups were examined using one-way analysis of variance (ANOVA) followed by the Tukey’s test for multiple comparisons. Categorical variables were compared using Chi square test. Correlations were analyzed using the Pearson test. The statistical evaluation of the data was performed using Graph Pad Prism 4.03 (GraphPad Software, San Diego, California, USA). P values < 0.05 were considered statistically significant.
Results
Long non-coding RNA CCHE1 (lncRNA-CCHE1) relative expression and its correlation with demographic and clinicopathological features of CRC patients
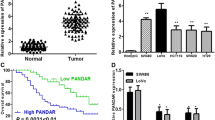
LncRNA-CCHE1 expression levels were assessed in colon cancer and adjacent non-cancerous tissues from 60 CRC patients using quantitative real-time PCR. Our data revealed that lncRNA-CCHE1 expression levels were significantly upregulated in colon cancer specimens compared with the matched adjacent normal colon tissues (p < 0.0001). Moreover, lncRNA-CCHE1 expression levels were significantly increased in patients with Grade II CRC compared to those with Grade I tumor; and in patients with Grade III CRC compared to those with Grade I and Grade II tumor (p < 0.0001). Likewise, lncRNA-CCHE1 expression levels were markedly upregulated in CRC patients with positive lymph node involvement compared to those with negative lymph node involvement (p < 0.0001). These data are illustrated in Figs. 1, 2.
a Long non-coding RNA CCHE1 (lncRNA-CCHE1) relative expression levels were significantly higher in CRC patients with advanced tumor grade. b Long non-coding RNA CCHE1 (lncRNA-CCHE1) relative expression levels were significantly higher in CRC patients with positive lymph nodes involvement. *P was considered significant at < 0.05
To explore whether the increased expression of lncRNA-CCHE1 was associated with the patients’ demographic and clinicopathological features such as age, gender, tumor location, tumor size, tumor histologic type, Dukes’Tumor stage, tumor histological type, tumor grade, lymph node involvement as well as vascular and perineural invasion, we classified the patients into two groups on the basis of their qRT-PCR results for lncRNA-CCHE1: high expression group where the relative expression level of lncRNA-CCHE1 is fivefold than para-carcinoma tissue, and low expression group where the relative expression level of lncRNA-CCHE1 is less than fivefold than para-carcinoma tissue.
As depicted in Table 1, that increased lncRNA-CCHE1 expression was significantly correlated with larger tumor size (p = 0.010), advanced Dukes ‘tumor stage (p = 0.017), advanced tumor grade or less differentiated histology (p = 0.019), positive lymph node involvement (p = 0.003), as well as positive vascular invasion (p = 0.0003). However, no significant difference was observed in terms of the patient age, gender, tumor localization, tumor histologic type, and tumor perineural invasion. These data suggest that lncRNA-CCHE1 upregulation is strongly associated with the clinical progression of human CRC.
Phosphorylated extracellular signal regulated kinase1/2 (p-ERK1/2) and cyclin D1 immunoassay
Our results demonstrated that both p-ERK1/2 and cyclin D1 levels were significantly elevated in colon cancer specimens compared with the matched adjacent non-cancerous (control) tissues from 60 CRC patients (p < 0.0001).These data are demonstrated in Fig. 3.
a Phosphorylated extracellular signal regulated kinase1/2 (p-ERK1/2) levels (pg/mg protein) in colon cancer and adjacent non-cancerous tissues (control) from CRC patients (n = 60). b Cyclin D1 levels (pg/mg protein) in colon cancer and adjacent non-cancerous tissues (control) from CRC patients (n = 60). Values are expressed as mean ± SD. *P was considered significant at < 0.05
Immunohistochemical staining for COX-2 expression in colorectal carcinoma
Positive immunoreactivity to COX-2 was detected as a brown cytoplasmic staining to tumor cells and not to the normal mucosal cells. Our data revealed that COX-2 expression was positive (3–7 of the final staining score) in 70% (42/60) and negative (0–2) in 30% (18/60) of the studied cases. Regarding the tumor histological type; 73.3% of conventional adenocarcinoma and 60% of mucinous adenocarcinoma cases were positive. There was no significant association between COX-2 expression and patient age (P = 0.41), gender (P = 0.47), tumor location (P = 0.89), tumor histological type (P = 0.32), and perineural invasion (P = 0.37). On the other hand, COX-2 expression showed significant differences with various pathological characteristics, including tumor size (P = 0.035), Dukes’ tumor stage (P = 0.0001), tumor differentiation (P ≤ 0.0001), as well as lymph node (P = 0.003) and vascular invasion (P = 0.03).
Of note, COX-2 expression showed positive association with lncRNA-CCHE1 expression, where the frequency of positive COX-2 expression was significantly higher in CRC cases with high lncRNA-CCHE1 expression (84.4) than in those with low lncRNA-CCHE1 expression (53.6%, P = 0.009). These data are demonstrated in Table 2 and Fig. 4a–c.
a–c Immunohistochemical staining for COX-2 expression in colorectal adenocarcinoma. a Weak cytoplasmic positivity (× 400); b moderate cytoplasmic positivity (× 200); c strong cytoplasmic positivity (× 200). d–f Immunohistochemical staining for PCNA expression in colorectal adenocarcinoma. d Negative nuclear immunostaining (× 100); e moderate nuclear immunostaining (× 200); f strong nuclear immunostaining (× 200)
Immunohistochemical staining for Proliferating cell nuclear antigen (PCNA) expression in colorectal carcinoma
Immunoreactivity to PCNA appeared as diffuse or granular nuclear staining. PCNA was scored according to the percentage of positive nuclei. Our data revealed that PCNA expression was positive (≥ 25% of the final staining score) in 76.6% (46/60) and negative (< 25%) in 23.3% (14/60) of the studied cases. Among the positive cases; 73.9% had conventional adenocarcinoma type, while 26.1% had mucinous adenocarcinoma type. There was no significant association between PCNA expression and patient age (P = 0.58), gender (P = 0.66), tumor location (P = 0.85), tumor histological type (P = 0.72), and perineural invasion (P = 0.88). Alternatively, PCNA expression showed significant differences with various pathological characteristics, including tumor size (P = 0.001), Dukes’ tumor stage (P = 0.004), tumor differentiation (P = < 0.0001), as well as lymph node (P = 0.004) and vascular invasion (P = 0.006).
Noteworthy, PCNA expression showed a significant positive association with lncRNA-CCHE1 expression, where the frequency of positive PCNA expression was markedly higher in CRC cases with high lncRNA-CCHE1 expression (93.8%) than in those with low lncRNA-CCHE1 expression (57.1%, P = 0.0008). These data are demonstrated in Table 2 and Fig. 4d–f.
Associations between some studied parameters in colon cancer and adjacent non-cancerous tissues from CRC patients
The correlations between some studied parameters in colon cancer and adjacent non-cancerous tissues from CRC patients (n = 60) were performed using Pearson’s correlation, where a significant positive correlations were revealed among the relative expression levels of lncRNA-CCHE1, and both tissue p-ERK1/2(r = 0.543, p < 0.0001) and cyclin D1 levels (r = 0.597, p < 0.0001). Moreover, another positive correlation was also observed between p-ERK1/2 and cyclin D1 tissue levels(r = 601, p < 0.0001). These data are summarized in Table 3. Nevertheless, as illustrated in Fig. 5, lncRNA-CCHE1 gene expression was significantly increased in CRC tissue specimens exhibiting positive COX-2 immunohistochemical staining compared to those with negative COX-2 staining (p < 0.0001). A similar pattern is observed between lncRNA-CCHE1 expression and PCNA immunohistochemical staining.
Discussion
Colorectal carcinoma (CRC) remains a major health concern worldwide; deciphering the molecular mechanisms underlying CRC pathogenesis is urgently warranted in order to provide tailored diagnostic and therapeutic strategies [21].
To the best of our knowledge, our study is the first to investigate the expression of the novel long noncoding RNA CCHE1 in CRC tissues; and to scrutinize its potential correlation with ERK/COX-2 pathway, some cell cycle and proliferation markers, as well as clinicopathological characteristics.
Our data revealed that that the expression of the lncRNA-CCHE1 was significantly up-regulated in CRC tissues as compared to their adjacent normal tissues. Next, we analyzed the correlation between lncRNA-CCHE1 expression and the clinicopathological characteristics of our cohort of colorectal cancer patients. Our results depicted that high lncRNA-CCHE1 expression was positively associated with larger tumor size, less differentiated histology, advanced dukes’ stage, positive lymph node involvement and vascular invasion; lending credence to the notion that lncRNA-CCHE1 might contribute to CRC pathogenesis and could potentially be exploited as a novel unfavorable prognostic biomarker for CRC progression monitoring.
This finding extends recent data demonstrating that lncRNA-CCHE1 is upregulated in certain types of cancer. For instance, lncRNA CCHE1 expression was upregulated in hepatocellular carcinoma [22] and cervical cancer tissues [8] and significantly correlated with tumor size, advanced tumor stage and poor overall survival; implying that it might enhance HCC and cervical cancer progression and could serve as biomarker for poor prognosis [22, 23]. Well in line, Zhan and colleagues [15] provided evidence that lncRNA CCHE1 overexpression promotes tumorigenesis and indicates poor prognosis in urothelial bladder carcinoma.
Likewise, Xu et al. [24] found that lncRNA CCHE1 up-regulation could promote cell proliferative ability and colony formation and inhibit cell apoptosis in gastric cancer cell lines. Moreover, Liao et al. [25] noted that lncRNA CCHE1 was highly expressed in the non small cell lung cancer (NSCLC) tissue, and its expression was closely related to the tumor size and poor survival. They also showed that lncRNA CCHE1 could promote the proliferation, metastasis, and invasion of NSCLC cell line. Collectively, these data allow us to speculate an oncogenic role for lncRNA-CCHE1 in promoting cancer cell proliferation and invasion.
To preliminarily explore the molecular mechanism through which lncRNA-CCHE1 could affect the biological cellular activities and thus contribute to CRC pathogenesis, an attempt was further made to assess the CRC tissue levels of p-ERK1/2, and its downstream target COX-2, and to examine their correlation with lncRNA-CCHE1 expression.
Our data unsurprisingly demonstrates that p-ERK1/2 levels were markedly elevated in CRC tissues compared to adjacent normal tissues. These data are essentially consistent with previous studies showing that ERK signaling is frequently dysregulated in multiple human cancer types [26,27,28,29]. Activated ERK 1/2 was found to regulate various cellular processes such as cell proliferation, differentiation, epithelial–mesenchymal transition, migration and senescence in a cell context-dependant manner [27].
Furthermore, our results demonstrated that COX-2 was overexpressed in CRC tissues, and its high expression was significantly associated with less differentiated histology, advanced dukes’ tumor stage, supporting the hypothesis that COX-2 up-regulation is a key step in colorectal carcinogenesis being not only associated with the development of colorectal carcinoma, but also played a main role in its progression.
In agreement with our finding, Elzagheid et al. [30] have also reported that COX-2 expression displayed a significant correlation with tumor stage and hence is assumed to be a negative prognostic factor in CRC patients. Also, Wu and Sun [31] have identified COX-2 as an important biomarker for invasion and metastasis of colorectal cancer.
These findings are biologically plausible since prostanoids, whose synthesis is catalyzed by COX-2, are crucial in the regulation of cell proliferation, angiogenesis, inflammation, tumor cell invasiveness and apoptosis resistance [32], they also contributes to immune evasion and resistance to cancer immunotherapy which may all contribute to CRC development and progression [29].
Intriguingly, both p-ERK1/2 levels and COX-2 expression showed a significant positive correlation with lncRNA-CCHE1 expression levels. This finding gives rise to the thought that the potential oncogenic role for lncRNA-CCHE1 in CRC could be mediated, at least in part, by regulating ERK/COX-2 pathway.
In line with this premise, Peng et al. [22] reported that CCHE1 knockdown significantly impaired HCC cell growth in vitro, secondary to decreased ERK and MAPK phosphorylation and therefore ERK/MAPK pathway inhibition. Likewise, Liao et al. [25] further corroborated this notion by demonstrating that suppression of lncRNA-CCHE1 expression could decrease ERK/MAPK expression and phosphorylation in NSCLC cell line and hence concluding that lncRNA-CCHE1 could promote the proliferation, metastasis, and invasion of NSCLC cell line via increasing the activating ERK/MAPK signaling pathway. However, the direct link between lncRNA-CCHE1 and the ERK/COX-2 pathway needs to be further investigated.
Since uncontrolled cell proliferation and disturbed cell cycle kinetics are the hallmarks of cancer [33], we thus sought to assess the levels of cyclin-D1and PCNA expression in CRC as markers for cell proliferation and to inspect their correlation with lncRNA-CCHE1 expression.
Our data revealed that cyclin-D1 levels, a key regulator of the G1 progression step within the cell cycle, were markedly elevated in CRC tissues compared to adjacent normal tissues. In this sense, it has been reported that cyclin D1is overexpressed in many human cancers where it functions as an oncogene promoting carcinogenesis and cancer cell cycle progression [34]. Moreover, a recent metaanalysis of 22 studies comprising 4150 CRC patients attested that Cyclin D1 is an unfavorable prognostic factor in CRC patients, since its over-expression was associated with poor clinical outcome and some clinicopathological features such as age, advanced clinical stage and distant metastasis [35].
Of note, our data displayed that cyclin-D1 and p- ERK ½ levels in colonic tissues were significantly correlated; underscoring the concept that cyclin D1 expression is positively regulated by MEK/ERK signaling, and sustained ERK activation is required for cyclin D1 continued expression [36].
PCNA is a cell cycle-associated protein, and its highest expression occurs in late G1 and S phases of the cell cycle [37]. Our results demonstrated that PCNA was over-expressed in CRC tissues, and its high expression was significantly associated with less differentiated histology, advanced dukes’ tumor stage.
Importantly, we observed that lncRNA-CCHE1 expression significantly correlated with the expression PCNA and cyclin-D1; proving the correlation between lncRNA-CCHE1 expression and cell proliferation activity, as well as bolstering the hypothesis that lncRNA-CCHE1 might be a potential biomarker for CRC because proliferation activity is a reliable marker for cancer progression.
Along this line, Yang et al. [8] attested a direct liaison between lncRNA-CCHE1and PCNA by reporting that lncRNA-CCHE1 overexpression significantly increased the mRNA and protein level of PCNA, probably through its interaction with PCNA messenger RNA, in vitro. Concomitantly, Zhan et al. [15] verified that lncRNA-CCHE1upregulates PCNA expression and serves as a key regulator in urothelial bladder carcinoma development.
The present study has some limitations that merit mentioning. First, it is a preliminary in vivo study; therefore, in vitro experiments on CRC cell lines are needed to confirm the direct effects of both lncRNA-CCHE1overexpression and silencing with specific siRNA on the investigated parameters. Also, as the number of patients is relatively small, our findings should be verified using larger scaled studies.
In conclusion, our study demonstrated, for the first time, that lncRNA-CCHE1expression was upregulated in CRC tissues, and was significantly associated with p-ERK1/2 and COX-2 expression, some cell proliferation markers, as well as to bad prognostic factors. These findings signify that lncRNA-CCHE1 is a key oncogene possibly involved in CRC pathogenesis by modulating ERK/COX-2 pathway and cell proliferation activity. Our study also provides a rationale for potential use of lncRNA-CCHE1 as a novel prognostic marker for monitoring CRC progression, and opens the door for the development of lncRNA-CCHE1-directed therapeutic strategies for CRC patients.
Abbreviations
- CRC:
-
Colorectal cancer
- lncRNA-CCHE1:
-
Cervical carcinoma high-expressed lncRNA 1
- HOTAIR:
-
Hox transcript antisense intergenic RNA
- MALAT1:
-
Metastasis-associated lung adenocarcinoma transcript 1
- BANCR:
-
BRAF-activated non-protein coding RNA
- HULC:
-
Highly upregulated in liver cancer
- ERK1/2:
-
Extracellular signal regulated kinase1/2
- COX-2:
-
Cyclooxygenase-2
- PCNA:
-
Proliferating cell nuclear antigen
References
Siegel RL et al (2017) Colorectal cancer statistics, 2017. CA Cancer J Clin 67(3):177–193
Tariq K, Ghias K (2016) Colorectal cancer carcinogenesis: a review of mechanisms. Cancer Biol Med 13(1):120
Schmitt AM, Chang HY (2016) Long noncoding RNAs in cancer pathways. Cancer Cell 29(4):452–463
Kashi K et al (2016) Discovery and functional analysis of lncRNAs: methodologies to investigate an uncharacterized transcriptome. Biochim Biophys Acta 1859(1):3–15
Xie X et al (2016) Long non-coding RNAs in colorectal cancer. Oncotarget 7(5):5226–5239
Ragusa M et al (2015) Non-coding landscapes of colorectal cancer. World J Gastroenterol 21(41):11709–11739
Deng H et al (2017) Long non-coding RNAs: new biomarkers for prognosis and diagnosis of colon cancer. Tumor Biol 39(6):1010428317706332
Yang M et al (2015) Long noncoding RNA CCHE1 promotes cervical cancer cell proliferation via upregulating PCNA. Tumor Biol 36(10):7615–7622
Liu B, Qu L, Yan S (2015) Cyclooxygenase-2 promotes tumor growth and suppresses tumor immunity. Cancer Cell Int 15(1):106
Bhattacharyya S et al (2016), Tenascin-C drives persistence of organ fibrosis. Nat Commun 7:11703
Sun Y et al (2015) Signaling pathway of MAPK/ERK in cell proliferation, differentiation, migration, senescence and apoptosis. J Recept Signal Transduct 35(6):600–604
Lim S, Kaldis P (2013) Cdks, cyclins and CKIs: roles beyond cell cycle regulation. Development 140(15):3079–3093
Xu MD, Qi P, Du X (2014) Long non-coding RNAs in colorectal cancer: implications for pathogenesis and clinical application. Mod Pathol 27(10):1310–1320
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72(1–2):248–254
Zhan Y et al (2017) Increased expression of long non-coding RNA CCEPR is associated with poor prognosis and promotes tumorigenesis in urothelial bladder carcinoma. Oncotarget 8(27):44326–44334
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 – ∆∆CT method. Methods 25(4):402–408
Broders AC (1925) The grading of carcinoma. Minn Med 8(726):1730–1925
Dukes CE (1949) The surgical pathology of rectal cancer. J Clin Pathol 2(2):95–98
Remmele W, Stegner HE (1987) Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathologe 8(3):138–140
Sun XF et al (1996) Proliferating cell nuclear antigen (PCNA) in relation to ras, c-erbB-2,p53, clinico-pathological variables and prognosis in colorectal adenocarcinoma. Int J Cancer 69(1):5–8
Grady WM, Markowitz SD (2015) The molecular pathogenesis of colorectal cancer and its potential application to colorectal cancer screening. Dig Dis Sci 60(3):762–772
Peng W, Fan H (2016) Long noncoding RNA CCHE1 indicates a poor prognosis of hepatocellular carcinoma and promotes carcinogenesis via activation of the ERK/MAPK pathway. Biomed Pharmacother 83:450–455
Chen Y et al (2017) Long non-coding RNA CCHE1 overexpression predicts a poor prognosis for cervical cancer. Eur Rev Med Pharmacol Sci 21(3):479–483
Xu G et al (2018) LncRNA CCHE1 in the proliferation and apoptosis of gastric cancer cells. Eur Rev Med Pharmacol Sci 22(9):2631–2637
Liao Y et al (2018) lncRNA CCHE1 increased proliferation, metastasis and invasion of non-small lung cancer cells and predicted poor survival in non-small lung cancer patients. Eur Rev Med Pharmacol Sci 22(6):1686–1692
Scrima M et al (2017) Aberrant signaling through the HER2-ERK1/2 pathway is predictive of reduced disease-free and overall survival in early stage non-small cell lung cancer (NSCLC) patients. J Cancer 8(2):227
Bai L et al (2015) ERK1/2 promoted proliferation and inhibited apoptosis of human cervical cancer cells and regulated the expression of c-Fos and c-Jun proteins. Med Oncol 32(3):57
Zhao L et al (2015) Benzidine induces epithelial-mesenchymal transition in human uroepithelial cells through ERK1/2 pathway. Biochem Biophys Res Commun 459(4):643–649
Ding G et al (2015) Over-expression of lipocalin 2 promotes cell migration and invasion through activating ERK signaling to increase SLUG expression in prostate cancer. Prostate 75(9):957–968
Elzagheid A et al (2013) High cyclooxygenase-2 expression is associated with advanced stages in colorectal cancer. Anticancer Res 33(8):3137–3143
Wu QB, Sun GP (2015) Expression of COX-2 and HER-2 in colorectal cancer and their correlation. World J Gastroenterol 21(20):6206–6214
Rizzo MT (2011) Cyclooxygenase-2 in oncogenesis. Clin Chim Acta 412(9–10):671–687
Barnum KJ, O’Connell MJ (2014) Cell cycle regulation by checkpoints. Cell Cycle Control 1170:29–40
Qie S, Diehl JA (2016) Cyclin D1, cancer progression, and opportunities in cancer treatment. J Mol Med 94(12):1313–1326
Li Y et al (2014) Prognostic significance of cyclin D1 expression in colorectal cancer: a meta-analysis of observational studies. PLoS ONE 9(4):e94508
Wang HY et al (2014) HBx protein promotes oval cell proliferation by up-regulation of cyclin D1 via activation of the MEK/ERK and PI3K/Akt pathways. Int J Mol Sci 15(3):3507–3518
Han DP et al (2012) Polo-like kinase 1 is overexpressed in colorectal cancer and participates in the migration and invasion of colorectal cancer cells. Med Sci Monit 18(6):Br237–B46
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Gaballah, H.H., Gaber, R.A., Elrashidy, M.A. et al. Expression of long non-coding RNA CCHE1 in colorectal carcinoma: correlations with clinicopathological features and ERK/COX-2 pathway. Mol Biol Rep 46, 657–667 (2019). https://doi.org/10.1007/s11033-018-4521-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-018-4521-0