Abstract
An important alternative source of fish oil is its production by plants through metabolic engineering. To produce eicosapentaenoic acid (EPA, 20:5n-3) in peanut through the alternative Δ8-pathway, a plant expression vector containing five heterologous genes driven by the constitutive 35S promoter respectively, namely, ∆9-elongase (Isochrysis galbana), ∆8-desaturase (Euglena gracilis), ∆5-desaturase (Mortierella alpina), ∆15-desaturase (Arabidopsis thaliana) and ∆17-desaturase (Phytophthora infestans) were transferred into peanut through Agrobacterium-mediated transformation method. The gas chromatography results indicated that the average content of EPA in the leaves of the transgenic lines was 0.68%, and the highest accumulation of EPA in an individual line reached 0.84%. This finding indicates that it is feasible to synthesize EPA in peanut through metabolic engineering and lays the foundations for the production of very-long-chain polyunsaturated fatty acids (VLCPUFAs) in peanut seeds.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Eicosapentaenoic acid (EPA, 20:5n-3) is an important component of fish oil and plays a beneficial role in many processes, including inflammation, immunity, blood pressure control, platelet aggregation, and gene regulation [1,2,3,4]. Therefore, it is important to obtain a sufficient amount of EPA from the daily diet because the human body has a very limited capacity to synthesize this very-long-chain polyunsaturated fatty acid (VLCPUFA) [5, 6]. Currently, the major dietary source of VLCPUFAs is oily marine fish; however, the declining fish stocks as a result of over-fishing and ever-increasing serious environmental pollution forced us to search for novel alternative sources.
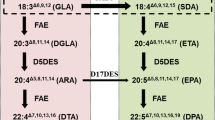
An appealing alternative source of fish oil production is higher plants with the capacity to synthesize these fatty acids effectively through metabolic engineering. There are two main aerobic biosynthetic pathways (the conventional Δ6-pathway and the alternative Δ8-pathway) for the biosynthesis of VLCPUFAs (Fig. 1). In the conventional Δ6-pathway, arachidonic acid (AA, 20:4n-6) and EPA are synthesized from two C18 fatty acids, linoleic acid (LA, 18:2n-6) and α-linolenic acid (ALA, 18:3n-3) by Δ6 desaturation, Δ6 elongation and Δ5 desaturation. In the alternative Δ8-pathway, the elongation and desaturation steps are inverted with respect to the traditional Δ6-pathway, wherein LA and ALA are first elongated to yield eicosadienoic acid (EDA, 20:2n-6) and eicosatrienoic acid (ETrA, 20:3n-3) by a Δ9 elongation, and then desaturated by two consecutive steps catalysted by a Δ8-desaturase and a Δ5-desaturase respectively to finally produce AA and EPA. Compared with the Δ6-pathway, the alternative Δ8-pathway mitigates the frequency of acyl exchanges between the phosphatidylcholine (PC) and acyl-CoA pool; thus, the alternative Δ8-pathway may be more efficient to synthesize EPA [7, 8]. During the last 10 years, various transgenic plants that produce VLCPUFAs through the Δ6-pathway have been obtained, and these plants include Linum usitatissimum (EPA, 0.8%) [9], Glycine max (EPA, 13.3%) [10], G. max (EPA, 19.6%) [10], G. max (EPA, 5.2%; DHA, 3.3%) [11], Brassica juncea (EPA, 8.1%; DHA, 0.2%) [12], Brassica carinata (EPA, 20.4%) [13], Nicotiana benthamiana (EPA, 10.7%) [14], Arabidopsis thaliana (EPA, 1.8%; DHA, 15.1%) [15], Camelina sativa (EPA, 3.3%; DHA, 12.4%), and C. sativa (EPA, 31% and EPA, 12%; DHA, 14%) [16]. The conventional Δ6-pathway has been studied earlier, but there was no distinctive progress. The first examples of the utilization of the Δ8-pathway for VLCPUFA synthesis in plants have been reported by Qi et al. [17]; EPA was accumulated to a significant level of 3% of total fatty acids in leaf tissues of A. thaliana. In our recent studies, the level of EPA in the leaves was up to 5.0% in transgenic cotton through expression of four genes under the control of the constitutive 35S promoter via the Δ8-pathway [18]. So far, the highest accumulation of EPA in transgenic plants was up to 14.6% via the ∆8-pathway, which was obtained in the seeds of an oilseed crop, C. sativa [19]. However, there have been few other examples reported that production of EPA in transgenic plants through Δ8-pathway except above studies until now.
Schematic representation of the EPA biosynthetic pathway involving the Δ8-pathway. LA and ALA can first be elongated by I. galbana Δ9-elongase and then by Euglena gracilis Δ8-desaturase and then further desaturated by Mortierella alpine Δ5-desaturase to produce AA and EPA, respectively. The ω6 fatty acids LA and AA can be converted to ALA and EPA by the ω3 desaturases A. thaliana Δ15 and Phytophthora infestans Δ17 desaturase, respectively
Peanut is one of the most important oil crops in the world and its leaves and stalks are good sources of livestock feed. It has a high average oil content of 50%, depending on the variety and agronomic conditions [20]. At present, the genetic transformation of peanut has been achieved through both the Agrobacterium-mediated transformation method using peanut seedling leaves or seed leaf as explants [21] and through the biolistic-mediated transformation method using embryonic callus as the transformation material [22]. However, both the Agrobacterium-mediated transformation method and the biolistic-mediated transformation method have the problems of low transformation and regeneration efficiencies as well as poor repeatability [23,24,25,26]. These factors limit the development and utilization of peanut as a “bioreactor” and the use of transgenic technology for trait improvement.
In this study, both a regeneration system and the Agrobacterium tumefaciens-mediated genetic transformation system were optimized using young leaves of peanut as explants. The alternative Δ8-pathway for the synthesis of EPA in peanut was fully reconstituted using the constitutive 35S promoter to drive the expression of five independent genes, namely, ∆9 elongase (Isochrysis galbana), ∆8-desaturase (Euglena gracilis), ∆5-desaturase (Mortierella alpina), ∆15-desaturase (A. thaliana) and ∆17-desaturase (Phytophthora infestans) respectively, which were transferred into peanut through the Agrobacterium-mediated transformation method. EPA was successfully produced in peanut leaves utilizing LA (18:2Δ9, 12) and ALA (18:3Δ9, 12, 15) as the primary substrates which are found at high concentrations in the leaves.
Materials and methods
Plant materials
Three peanut cultivars were used: Shanhua 7, Luhua 8 and Huayu 9616. These peanut cultivars were carefully selected and sown in the field at the farm station of the Crops Research Institute, Shandong Agricultural University, Taian, China.
Sterilization and culture establishment
The seeds were washed with tap water, surface-sterilized in 70% ethanol (v/v) for 1 min, treated with 0.1% HgCl2 (w/v) for 15 min, and rinsed five times in sterile distilled water. The cotyledons were then cut into two halves, and the seed coats were removed and cultured on MSB5 basal medium (MS salts and B5 vitamins). The young leaves, which emerged after 4 days, were cut into 2 × 2 mm pieces and then transferred to MSB5 basal medium supplemented with 6-BA and NAA to induce bud formation. After 4–5 weeks, when the length of the buds was approximately 1 cm, they were transferred to MSB5 basal medium supplemented with 6-BA for shoot elongation and transferred to MSB5 basal medium for rooting under the same conditions. All growth media were supplemented with 3% sucrose and 0.6% (w/v) agar. The cultures were incubated at 25 ± 2 °C under a 16-h photoperiod with cool white fluorescent lights (80 l mol/m2/s).
EPA expression vector and pCambia2300GFP vector construction
Using the multiple gene expression vector construction technology developed in our laboratory [27] to construct a plant binary vector containing five genes, we first transformed each gene from the PUC-T Simple cloning vector into the pAUX2 auxiliary vector (–XbaI–35S–MCS–Tnos–AvrII–XbaI–), which contains a 35S promoter, a multi-cloning site (MCS), and a Nos terminator (Tnos). Each target gene was subcloned into the MCS of the vector to generate an individual expression cassette. Because XbaI (T/CTAGA) and AvrII (C/CTAGG) can produce compatible cohesive ends, the five expression cassettes were cascaded together through repeated digestion and ligation with XbaI and AvrII. After these steps, a single large DNA fragment that contained the five gene cassettes was digested from this auxiliary vector with XbaI and ligated into the pCambia2300 plant binary vector pre-digested with XbaI to obtain a plant transformation vector (Fig. 2).
The pCambia2300GFP vector was constructed via the pCambia2300 plant expression vector, which was reformed by adding an expression EC box.
Agrobacterium-mediated peanut transformation
The EPA expression vector and the pCambia2300GFP vector were introduced into the A. tumefaciens strain GV3101. The kanamycin-resistance clone that contains all five genes or GFP was cultured to an OD600 of 0.8–1.0. The explants were incubated in the Agrobacterium suspension for 10 min. The leaf segments were then briefly blotted onto filter paper, placed onto MSB5 medium supplemented with 6.0 mg/l BA and 1.0 mg/l NAA and cultured in the dark at 22 °C for 3 days for co-culture. The cultures were then washed with sterile distilled water, briefly blotted onto filter paper and transferred into the resting medium (MSB5 medium supplemented with 6.0 mg/l 6-BA and 1.0 mg/l NAA, 500 mg/l cefotaxime, and 50 mg/l kanamycin).
DNA extraction and polymerase chain reaction (PCR)
Genomic DNA samples were obtained from plants using CTAB and were analyzed by PCR amplification using primers for Δ9-Elo, Δ8-Des, Δ5-Des, Δ15-Des and Δ17-Des (Table 1). The PCR amplification conditions consisted of pre-denaturation at 94 °C for 3 min, 40 cycles of denaturation at 94 °C for 1 min, annealing at an appropriate temperature and extension at 72 °C for 1 min, and a final extension at 72 °C for 10 min.
Fatty acid analysis
The total fatty acids from peanut were extracted and transmethylated with methanolic HCl according to a previously described procedure. The fatty acid methyl esters (FAMEs) were analyzed by gas chromatography (GC) on a 25 m × 0.25 mm, 0.20 mm, fused silica CP-Wax 52CB capillary column (Chrompack UK Ltd, London UK) using a split/splitless injector (230 °C, split ratio = 50:1) and a flame ionization detector (300 °C). After exposure to an initial temperature of 170 °C for 3 min, the column temperature was increased at a rate of 4 °C/min–220 °C and then maintained at 220 °C for 15 min. Hydrogen was used as the carrier gas at an initial flow rate of 1 ml/min. The FAMEs were identified through comparisons of their retention times with those of known standards. Their identities were further validated by GC-mass spectrometry (GC-MS) as previously described.
Results
Optimization of peanut genetic transformation system mediated by A. tumefaciens
In order to produce EPA in peanut by using the Agrobacterium-mediated transformation method, the regeneration system of peanut leaves was firstly optimized. Because shoot differentiation of callus is crucial for the establishment of a peanut regeneration system, and in which, the hormone concentration and peanut genotype have an important effect on shoot induction [21]. So, we screened the peanut genotypes and detected a proper hormone concentration for the shoot induction using 4 day-old young leaves derived from Shanhua 7, Luhua 8 and Huayu 9616 as explants, which were cultured on the induction culture medium supplemented with different concentration of 6-BA and NAA. After 4 weeks on the induction culture medium, the leaf sections became greenish, and buds were formed. The induction results indicated that the highest shoot induction rates for all three species were 63.8%, 70.9% and 51.8% on medium supplemented with 6.0 mg/l 6-BA and 1.0 mg/l NAA (Table S1and Fig. S1). In addition, the shoot induction rate of Luhua 8 was higher than that of the other species in almost all concentrations and reached 70.9% in the medium supplemented with 6.0 mg/l 6-BA and 1.0 mg/l NAA (Table S1 and Fig. S1). Thus, the hormone concentrations of 6.0 mg/l 6-BA and 1.0 mg/l NAA for the induction culture medium and Luhua 8 were chosen in our next research.
The infection time of Agrobacterium has an important effect on transformation rate. A long infection will lead to browning and little budding, whereas a short infection will reduce the conversion rate. The explants were incubated into the A. tumefaciens strain GV3101 suspension, which contains the pCambia2300GFP vector, for 5, 10, 15 and 20 min. After 5–7 weeks of cultivation, the number of fluorescent explants was counted. As shown in Table S2, the highest 8.16% conversion rate was obtained 10 min after infection, and the transformation rate reduced with an increase or decrease of infection duration. These results suggested that the Agrobacterium infection time has a significant influence on the conversion rate.
Kanamycin is commonly used to select for adventitious transformants. The explants of Luhua 8 were cultured on the induction culture medium containing 0, 50, 80, 100 and 150 mg/l kanamycin. No callus was found after culture in the medium containing 80 mg/l kanamycin, and 9% callus was regenerated with a kanamycin concentration of 50 mg/l (Table S3). In addition, no shoots were found after cultured with 50 mg/l and 80 mg/l kanamycin, indicating that the wild-type shoots cannot generate on medium containing 50 mg/l kanamycin even though there was a small amount of callus. Therefore, 50 mg/l kanamycin was used to screen transgenic transformants.
Regeneration of EPA transgenic plantlets
A peanut regeneration system was established using the young leaves of Luhua 8 as explants under the above-described optimal conditions. Subsequently, to produce EPA via the alternative Δ8-pathway, five independent genes under the control of the constitutive 35S promoter were transformed into peanut by Agrobacterium-mediated transformation method (Figs. 1, 2). The procedure for transgenic plantlet regeneration includes the whole tissue culture process as shown in Fig. 3a–i (Fig. 3a, b refer to the stages before and after the regeneration of the young leaves, and Fig. 3c refers to the pre-culture stage), after infection with A. tumefaciens, the explants were cultured on the selected medium for bud induction (Fig. 3d, e), and when the buds grew to 1 cm (Fig. 3e), they were transferred to the shoot elongation medium (Fig. 3f) and then to the rooting medium (Fig. 3g), where some coarse and lateral roots were observed. After the hardening-seedling step (Fig. 3h), the seedlings were transplanted to flowerpots (Fig. 3i), where the transgenic plants could flower and bear fruit.
Regenerative process of leaf material of peanut. a Seeds of mature peanut germination on shoot induction medium. b Regeneration of young leaves from seeds after 4 days of culture on medium. c Leaf segments of 2 × 2 mm cut from leaf explants. d Cluster buds formed in callus induction medium. e, f Elongation of cluster buds in shoot elongation medium. g Regenerated seedlings in rooting medium. h Rooted plantlet. i Transgenic seedling transplanted to a flowerpot
Identification of EPA transgenic lines by PCR
To identify the EPA transgenic lines in these transformants, the DNA from the putative transgenic plants was prepared and subjected to PCR amplification. Using gene-specific primers (Table 1), the target genes ∆9-elongase (I. galbana), ∆8-desaturase (E. gracilis), ∆5-desaturase (M. alpina), ∆15-desaturase (A. thaliana) and ∆17-desaturase (P. infestans), which are 559, 1095, 685, 1353 and 1086 bp in length, respectively, were amplified in the transgenic lines 5, 7, 10, 15 and 16 (Fig. 4). Therefore, these five lines were used for fatty acid analysis.
PCR analysis of resistant plant lines 5, 6, 7, 10, 15 and 16. Lane M: molecular weight marker; lane C: DNA from untransformed plant (negative control); lane P: pCAMBIA2300 plasmid DNA with △9, △8, △5, △15 and △17 genes (positive control); lane line 5–line 16: DNA from independently transformed plants
Fatty acid analysis of EPA transgenic lines by GC
To verify whether the target genes were expressed in the transgenic plants, the total fatty acids from leaves of the above-mentioned lines were extracted and analyzed by GC (Table 2; Fig. 5a, b). Compared with the fatty acids analysis of the wild type strain, two types of new fatty acids, namely, ETrA (20:3Δ11,14,17) and EPA (20:5Δ5,8,11,14,17), were found in the transgenic plants with mean contents of 1.24% and 0.68%, respectively. These finding indicated that it is feasible to produce EPA in peanut leaves. However, no 20:4(n-3) was found in the transgenic plants, which indicated that the enzyme activity of ∆5-desaturase may be higher than that of ∆8-desaturase. Therefore, almost all of the intermediate products were translated into final products. In addition, compared with the wild type, the fatty acid contents of 18:2(n-6) and 18:3(n-3) were decreased. Moreover, the total contents of ω6 VLCPUFAs were lower than those of ω3 VLCPUFAs, indicating that the heterologous genes ∆15-desaturase and ∆17-desaturase play a role in the transgenic plants.
Discussion
With the development of biotechnology, more and more nutrition enrichment transgenic crops with were produced, such as “Golden Rice”, “Purple Embryo Maize” and “Purple Endosperm Rice” [28,29,30]. The peanut seeds were rich in oil, especially oleic acid (OA, 18:1n-9) and LA, accounting for 80%, which are the precursors of EPA. The peanut is ideal organism to produce EPA as bioreactor, but no study has been attempted to produce EPA in peanut so far, although many examples for the synthesis of EPA in different varieties of plant via the conventional ∆6-pathway or the alternative Δ8-pathway have been reported. Our present study provides the first demonstration of the production of EPA in peanut through the alternative Δ8-pathway.
In the present study, protocols for efficient regeneration of multiple adventitious shoot buds from cotyledon explants of peanut has been reported [21, 24]. From these protocols, we learned that the hormone concentration and peanut genotype have an important effect on shoot induction. Thus, in this study, we first optimized the regeneration system of peanut using young leaves as explants. Our results showed that the highest bud induction rate was obtained with the medium supplemented with 1.0 mg/l NAA and 6.0 mg/l 6-BA. Therefore, 1.0 mg/l NAA and 6.0 mg/l 6-BA were added to the bud induction medium, and these may be the optimal concentrations for the induction of peanut buds of different genotypes. We used three peanut cultivars, namely Shanhua 7, Luhua 8 and Huayu 9616. The shoot induction rate of Luhua 8 was higher than those obtained with the other two species at almost all concentrations, and the highest rate obtained was 70.9%.
Using the young leaves of Luhua 8 as transgenic receptors, five heterologous genes under control by the constitutive 35S promoter respectively were introduced into peanut through Agrobacterium-mediated transformation method for the synthesis of EPA via the alternative Δ8-pathway. The target product could be detected in the leaves and the feasibility of producing EPA by peanut as a reactor could be verified in a short time. In addition, EPA could be produced through 35S promoter driving these genes expression in all parts of transgenic peanut which could be used as green feed for livestock. Then human can get EPA from these animals by ingesting their meat and milk, which avoids eating GM foods directly. The GC results showed that the average content of EPA in the leaves of the transgenic lines was 0.68%, and the highest accumulation of EPA in an individual line reached 0.84%. In addition, some new fatty acids, which are intermediates in the biosynthetic pathway, such as 20:3(n-3), 20:2(n-6) and 20:3(n-6), but not 20:4(n-3), were found in the GC profile of the transgenic plants. Because the five genes in the alternative Δ8-pathways for the synthesis of EPA were cascaded closely together in an independent expression cassette and were driven by the same promoter, they exhibited the same expression levels. In the ∆8-pathway, ∆9 elongase, which catalyzes the first reaction, is usually the rate-limiting enzyme, and the activities of other enzymes are relatively higher than that of this enzyme [31]. Thus, it is reasonable that no or few intermediate products were found in the leaves of the transgenic peanut, which is similar to the results obtained in some microalgae and fungi that are rich in EPA, such as P. infestans, where the EPA content is 15.2%, but the EPA synthetic intermediate contents are either very low or undetectable [32]. The production of fish oil with low levels of intermediates is markedly closer to the components of natural oils and is more likely to be accepted by the population.
In previous studies about the production of EPA via the alternative Δ8-pathway in transgenic plants, 3% EPA was obtained in the leaves of Arabidopsis which were sequential transformed with I. galbana Δ9-elongase, E. gracilis Δ8-desaturase and M. alpine Δ5-desaturase under the control of the constitutive 35S promoter [17], and the level of EPA was up to 5.0% in transgenic cotton [18] through expression of four enzyme genes include above three genes and an extra Arabidopsis Δ15-desaturase under the control of the constitutive 35S promoter. In this study, using the similar construct contained above four enzyme genes and an extra P. infestans Δ17-desaturase, the transgenic peanut leaves accumulated relatively low EPA level, and the highest accumulation of EPA was only 0.84%. Using the alternative Δ8-pathway for VLCPUFAs synthesis through expressing the different combinations of genes and promoters on the accumulation of EPA in an oilseed crop, C. sativa by Ruiz-Lopez et al. [19], the accumulation of EPA in the seeds was up to 14.6%, which was the highest accumulation of EPA in transgenic plants so far. For these differences of the EPA accumulation in different examples, a possible reason may be that different host plants have different lipid metabolic pathways and different relative levels of endogenous enzymes [33, 34]. Otherwise, the strong construct design obtained by engineering complex multi-gene pathways in a single construct has a marked effect on the expression of each gene [35]. The codon bias between the transgene donor and the host crop is one of the major contributing factors [36]. In other words, the judicious selection of host species and promoters, together with the inclusion of genes that enhance the basic VLCPUFA biosynthetic pathway, can greatly influence the production of EPA in plants.
In the process of the genetic modification of the fatty acid biosynthesis pathways in plants, the inefficiency of acyl exchanges between the PC and acyl-CoA pool is a major limiting factor that has a significant influence on the yield of VLCPUFAs [7, 8]. In theory, the existence form of fatty acid substrates acted on chain elongase and desaturase that involved in EPA synthesis pathway are different, in which elongase acts on the form of fatty acid-CoA and desaturase acts on the form of fatty acid-acyl [9]. The Δ6-pathway needs to undergo a chain desaturation–elongation–desaturation process, requiring two transformations of the substrate. However, the Δ8-pathway undergo a chain elongation–desaturation–desaturation process, so that there is only one transformation of the substrate. Therefore, the Δ8-pathway may be more conducive to the production of fish oil than the Δ6-pathways [17]. However, in this study, the low efficiency of producing fish oil using the Δ8-pathway may be related to the selection of promoters with the same sequence. It has been reported that the same promoter is liable to cause transgene silencing, which makes the efficiency of gene translation into protein low. It may also be related to the use of genes from lower organisms, whose codons are too different from the one used preferentially in peanut, which resulted in low yield of EPA [36]. In addition, compared with the transgenic plants with higher levels of EPA accumulation, the lower yield of EPA we studied here may be due to the number of transformed genes and the synthetic leaves for detection, but not seeds.
In practice, during the last ten years, various transgenic plants that produce VLCPUFAs have been generated via the conventional Δ6-pathway rather than the alternative Δ8-pathway, and the highest EPA content obtained in transgenic plants was 31% through the Δ6-pathway [16] and only 14.6% through the Δ8-pathway [19] respectively. The lower content of the target product in transgenic plants by using the alternative Δ8-pathway suggested that the production of EPA via the Δ8-pathway in transgenic plants may be more seriously limited by the source of substrates, the activity of enzymes and other elements compared with the Δ6-pathway. Thus, it is necessary to further understand how to best integrate the endogenous and transgene-derived metabolism.
Conclusions
In conclusion, this study provides the first demonstration of the possibility of synthesizing EPA through the Δ8-pathway in peanut and lays the foundation for the heterologous biosynthesis of fish oil through the Δ8-pathway in plants. However, the lower production of EPA obtained proves that many limited elements need to be resolved before the peanut can be used as a bioreactor for the alternative production of EPA. So, to further improve the EPA content in peanut, it should be necessary to choose highly active enzymes and favorable promoter, and to optimize the structure of genes in a more effective cascading order or increase the efficiency of the exogenous genes through codon usage optimization according to the host plant. Moreover, the expression of transcription factors, such as WRI1 and LEC1 [37], is vital in directing the carbon flux that enters the seed toward the synthesis of FAs. Thus, increasing the expression levels of endogenous transcription factors is another avenue that can be assessed in future studies to perfect our vision.
Abbreviations
- EPA:
-
Eicosapentaenoic acid
- VLCPUFAs:
-
Very-long-chain polyunsaturated fatty acids
- DHA:
-
Docosahexaenoic acid
- LA:
-
Linoleic acid
- ALA:
-
α-Linolenic acid
- 6-BA:
-
6-Benzyl aminopurine
- NAA:
-
1-Naphthylacetic acid
- MCS:
-
Multiple cloning site
- CTAB:
-
Cetyltrimethyl ammonium bromide
- Elo :
-
Elongase
- Des :
-
Desaturase
- GC:
-
Gas chromatography
- FAMEs:
-
Fatty acid methyl esters
- GC–MS:
-
Gas chromatograph–mass spectrometer
- ETrA:
-
Eicosatrienoic acid
References
Gill I, Valivety R (1997) Polyunsaturated fatty acids, part 1: occurrence, biological activities and applications. Trends Biotechnol 15:401–409
Lauritzen L, Hansen H, Jørgensen M, Michaelsen K (2001) The essentiality of long chain n-3 fatty acids in relation to development and function of the brain and retina. Prog Lipid Res 40:1–94
Thies F, Garry JM, Yaqoob P, Rerkasem K, Williams J, Shearman CP, Gallagher PJ, Calder PC, Grimble RF (2003) Association of n-3 polyunsaturated fatty acids with stability of atherosclerotic plaques: a randomised controlled trial. Lancet 361:477–485
Abeywardena P (2011) Role of ω3 long chain polyunsaturated fatty acids in reducing cardio-metabolic risk factors. Endocr Metab Immune Disord Drug Targets 11:232–246
Sprecher H (2000) Metabolism of highly unsaturated n-3 and n-6 fatty acids. Biochim Biophys Acta 1486:219–231
Pawlosky RJ, Hibbeln JR, Novotny JA, Salem N (2001) Physiological compartmental analysis of α-linolenic acid metabolism in adult humans. J Lipid Res 42:1257–1265
Snyder CL, Yurchenko OP, Siloto RM, Chen X, Liu Q, Mietkiewska E, Weselake RJ (2009) Acyltransferase action in the modification of seed oil biosynthesis. New Biotechnol 26:11–16
Yurchenko OP, Nykiforuk CL, Moloney MM, Ståhl U, Banaś A, Stymne S, Weselake RJ (2009) A 10-kDa acyl-CoA-binding protein (ACBP) from Brassica napus enhances acyl exchange between acyl-CoA and phosphatidylcholine. Plant Biotechnol J 7:602–610
Abbadi A, Domergue F, Bauer J, Napier J, Welti R, Zähringer U, Cirpus P, Heinz E (2004) Biosynthesis of very-long-chain polyunsaturated fatty acids in transgenic oilseeds: constraints on their accumulation. Plant Cell Online 16:2734–2748
Kinney A, Cahoon E, Damude H, Hitz W, Liu ZB, Kolar C (2004) Production of very long chain polyunsaturated fatty acids in oilseed plants, Google Patents 2004
Kinney A, Cahoon E, Damude H, Hitz W, Liu ZB, Kolar C (2011) Production of very long chain polyunsaturated fatty acids in oil seed plants, Google Patents 2011
Wu G, Truksa M, Datla N, Vrinten P, Bauer J, Zank T, Cirpus P, Heinz E, Qiu X (2005) Stepwise engineering to produce high yields of very long-chain polyunsaturated fatty acids in plants. Nat Biotechnol 23:1013–1017
Cheng B, Wu G, Vrinten P, Falk K, Bauer J, Qiu X (2010) Towards the production of high levels of eicosapentaenoic acid in transgenic plants: the effects of different host species, genes and promoters. Transgenic Res 19:221–229
Petrie JR, Shrestha P, Mansour MP, Nichols PD, Liu Q, Singh SP (2010) Metabolic engineering of omega-3 long-chain polyunsaturated fatty acids in plants using an acyl-CoA ∆6-desaturase with ω3-preference from the marine microalga Micromonas pusilla. Metab Eng 12:233–240
Petrie JR, Shrestha P, Zhou XR, Mansour MP, Liu Q, Belide S, Nichols PD, Singh SP (2012) Metabolic engineering plant seeds with fish oil-like levels of DHA. PLoS ONE 7:e49165
Ruiz-Lopez N, Haslam RP, Napier JA, Sayanova O (2014) Successful high-level accumulation of fish oil omega-3 long-chain polyunsaturated fatty acids in a transgenic oilseed crop. Plant J 77:198–208
Qi B, Fraser T, Mugford S, Dobson G, Sayanova O, Butler J, Napier JA, Stobart AK, Lazarus CM (2004) Production of very long chain polyunsaturated omega-3 and omega-6 fatty acids in plants. Nat Biotechnol 22:739–745
Liu J, Ma Y, Sun Q, Wu X, Li X, Sun M, Li Y, Li X, Qi B (2014) Production of very long chain polyunsaturated fatty acids in cotton. Acta Agron Sin 40:86–92
Ruiz-Lopez N, Haslam RP, Usher S, Napier JA, Sayanova O (2015) An alternative pathway for the effective production of the omega-3 long-chain polyunsaturates EPA and ETA in transgenic oilseeds. Plant Biotechnol J 13:1264–1275
Özcan M, Seven S (2003) Physıcal and chemıcal analysıs and fatty acıd composıtıon of peanut, peanut oıl and peanut butter from ÇOM and NC-7 cultıvars. Grasas Aceites 54:12–18
Sharma KK, Anjaiah V (2000) An efficient method for the production of transgenic plants of peanut (Arachis hypogaea L.) through Agrobacterium tumefaciens-mediated genetic transformation. Plant Sci 159:7–19
Klein TM, Wolf E, Wu R, Sanford J (1987) High-velocity microprojectiles for delivering nucleic acids into living cells. Nature 327:70–73
Rohini V, Rao KS (2001) Transformation of peanut (Arachis hypogaea L.) with tobacco chitinase gene: variable response of transformants to leaf spot disease. Plant Sci 160:889–898
Bhatnagar-Mathur P, Devi MJ, Reddy DS, Lavanya M, Vadez V, Serraj R, Yamaguchi-Shinozaki K, Sharma KK (2007) Stress-inducible expression of At DREB1A in transgenic peanut (Arachis hypogaea L.) increases transpiration efficiency under water-limiting conditions. Plant Cell Rep 26:2071–2082
Tiwari S, Mishra DK, Singh A, Singh P, Tuli R (2008) Expression of a synthetic crylEC gene for resistance against Spodoptera litura in transgenic peanut (Arachis hypogaea L.). Plant Cell Rep 27:1017–1025
Yang CY, Chen SY, Duan GC (2011) Transgenic peanut (Arachis hypogaea L.) expressing the urease subunit B gene of Helicobacter pylori. Curr Microbiol 63:387–391
Sun Q, Liu J, Li Y, Zhang Q, Shan S, Li X, Qi B (2013) Creation and validation of a widely applicable multiple gene transfer vector system for stable transformation in plant. Plant Mol Biol 83:391–404
Ye X, Al-Babili S, Klöti A, Zhang J, Lucca P, Beyer P, Potrykus I (2000) Engineering the provitamin A (β-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Science 287:303–305
Liu X, Yang W, Mu B, Li S, Li Y, Zhou X, Zhang C, Fan Y, Chen R (2018) Engineering of ‘Purple Embryo Maize’ with a multigene expression system derived from a bidirectional promoter and self-cleaving 2A peptides. Plant Biotechnol J 16:1107–1109
Zhu Q, Yu S, Zeng D, Liu H, Wang H, Yang Z, Xie X, Shen R, Tan J, Li H, Zhao X, Zhang Q, Chen Y, Guo J, Chen L, Liu YG (2017) Development of ‘‘Purple Endosperm Rice’’ by engineering anthocyanin biosynthesis in the endosperm with a high-efficiency transgene stacking system. Mol Plant 10:918–929
Pereira SL, Leonard AE, Mukerji P (2003) Recent advances in the study of fatty acid desaturases from animals and lower eukaryotes. Prostaglandins Leukot Essent Fatty Acids 68:97–106
Sun Q, Liu J, Zhang Q, Qing X, Dobson G, Li X, Qi B (2013) Characterization of three novel desaturases involved in the delta-6 desaturation pathways for polyunsaturated fatty acid biosynthesis from Phytophthora infestans. Appl Microbiol Biotechnol 97:7689–7697
Haslam RP, Ruiz-Lopez N, Eastmond P, Moloney M, Sayanova O, Napier JA (2013) The modification of plant oil composition via metabolic engineering-better nutrition by design. Plant Biotechnol J 11:157–168
Chen Y, Meesapyodsuk D, Qiu X (2014) Transgenic production of omega-3 very long chain polyunsaturated fatty acids in plants: accomplishment and challenge. Biocatal Agric Biotechnol 3:38–43
Petrie JR, Shrestha P, Belide S, Kennedy Y, Lester G, Liu Q, Divi UK, Mulder RJ, Mansour MP, Nichols PD (2014) Metabolic engineering Camelina sativa with fish oil-like levels of DHA. PLoS ONE 9:e85061
Xia F, Li X, Li X, Zheng D, Sun Q, Liu J, Li Y, Hua J, Qi B (2016) Elevation of the yields of very long chain polyunsaturated fatty acids via minimal codon optimization of two key biosynthetic enzymes. PLoS ONE 11:e0158103
Lu C, Napier JA, Clemente TE, Cahoon EB (2011) New frontiers in oilseed biotechnology: meeting the global demand for vegetable oils for food, feed, biofuel, and industrial applications. Curr Opin Biotechnol 22:252–259
Acknowledgements
This study was funded by the Shandong Province Peanut Seed Industry Project, the National Natural Science Foundation of China (31601336, 31271757) and the National Opening Project of State Key Laboratory of Crop Biology (2016KF09). The sequencing and assembly were performed by the Beijing Genomics Institute (BGI).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, C., Qing, X., Yu, M. et al. Production of eicosapentaenoic acid (EPA, 20:5n-3) in transgenic peanut (Arachis hypogaea L.) through the alternative Δ8-desaturase pathway. Mol Biol Rep 46, 333–342 (2019). https://doi.org/10.1007/s11033-018-4476-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-018-4476-1