Abstract
Bamboo is one of the important plant for pulp, paper and charcoal industries. After China, India is the second largest bamboo reserve in Asia. Around the globe, wide genetic diversity of bamboo is present which serves as the base for selection and improvement. DNA based molecular markers appears to be a striking substitute for systematic assessment of the genetic diversity in conservation and genetic improvement of plants. DNA based molecular markers such as RAPD and ISSR were used to assess the genetic diversity in 13 bamboo genotypes. Total 120 RAPD and 63 ISSR primers were tested, of which only 42 polymorphic primers (30 RAPD and 12 ISSR), gave reproducible amplification profile and were used in this study. 30 RAPD primers yielded total 645 amplified fragments, of which 623 were polymorphic, and 20.76 polymorphic bands per primer were observed across 13 genotypes. 12 ISSR primers produced 246 amplified fragments, of which 241 were polymorphic, and 20.08 polymorphic bands per primer was observed across 13 different genotypes. The Jaccard’s coefficient of RAPD, ISSR and pooled RAPD and ISSR dendrograms ranged from 0.26 to 0.83, 0.23 to 0.86 and 0.26 to 0.84 respectively. The present study found the large genetic diversity present between different elite genotypes of bamboo. Such investigation can deliver a well understanding of the available genotypes, which might be further exploited for the paper industry.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bamboo form the backbone of rural economy of Southeast Asian countries where it considered as the most important forest species. After China, India is the second biggest producer of bamboos in the world. India is generating four to six million tons of bamboos yearly, out of which two million tons are consumed in paper production. In India bamboo is commonly famous as “poor man’s wood” and considered as green gold. Apart from their application in paper production, it is widely used in household construction, furniture manufacture, flooring tiles, conservation of soil, food purposes and cattle food. Bamboo is a representative species, a member of five large subfamilies Arundinoideae, Pooideae, Panicoideae and Bambusoideae. Bamboo is perennial, giant, woody grass comprising of 60–70 genera with 1200–1500 species belonging to subfamily Bambusoideae [1]. Among which, 30 genera and 136 species are found in India.
Genetic destruction and extensive exploitation of bamboo species have made it compulsory not only for the conservation and protection of its germplasm [2, 3], but also to organize and illustrate them [4–6]. Classification of germplasm is a key link between the conservation and utilization of germplasm [7, 8]. To maintain and preserve the germplasm biodiversity, the exploration of bamboo resources and study of their native dissemination is necessary, which is documented to be limited till date [6–9].
Molecular markers have become an important part of most of the genetic diversity studies. Several DNA-based molecular markers, such as, inter simple sequence repeats (ISSR) markers, random amplified polymorphic DNA (RAPD), restriction fragment length polymorphism (RFLP), and amplified fragment length polymorphism (AFLP) have been used to explore genetic diversity. These molecular markers reveal the phylogenetic relationships among various genotypes, for their effective use in breeding, conservation and improvement. AFLP and RFLP used the radioactive labelling, expensive restriction enzyme, hence, appear inappropriate. On the other hand RAPD and ISSR markers, involved very less quantity of DNA without using radioactive labels, restriction enzyme, and are faster as well as simpler. RAPD has confirmed to be relatively effective in recognising genetic difference, and used for diversity evaluation and for finding new genotypes in many crops/plants [10–16]. ISSR has been revealed to deliver a potent, fast, reproducible, simple, and economical means to evaluate genetic variation and detect changes between narrowly related germplasm in various plant species [14, 17, 18]. Few investigations have been reported on phylogenetic and the genetic diversity of bamboo using DNA based marker [6, 9]. Considering the importance of work and potential of DNA based molecular marker based genetic diversity analysis, the present study was conducted to explore and depict the genetic diversity of Indian genotypes of bamboo using RAPD and ISSR markers.
Materials and methods
Genomic DNA isolation
DNA was isolated from 13 genotypes of bamboo (Table 1), by the original method described by Doyle and Doyle with few changes (viz., amount of leaf tissue, concentration of components of the buffer, and temperature) [19]. Fresh leaf tissues (0.2 g) were crushed in liquid nitrogen and transferred into a 2 ml pre-sterilised centrifuge tube. 0.5 ml of extraction buffer [100 mM tris–HCl, 0.5 M ethylenediaminetetraacetic acid (EDTA), 3.5 M NaCl, 3 % cetyltrimethyl ammonium bromide (CTAB), 1 % polyvinylpyrrolidone (PVP), and 0.2 M β-mercaptoethanol pH 8.0] was added in crushed sample and incubated at 65 °C for 60 min. After incubation samples were emulsified with an equal volume of chloroform: isoamyl alcohol (24:1) and supernatants were transferred into new pre-sterilised centrifuge tube and it was repeated twice. Precipitation of DNA was carried out using the mixture of chilled absolute alcohol and 3 M sodium acetate and by overnight incubation at −20 °C. Finally DNA was precipitated by chilled 80 % ethanol. The precipitated DNA was air dried and dissolved in 100 µl of Tris–EDTA (TE) buffer. The dissolved DNA was treated with RNase (2.25 U). Total pure DNA was qualified and quantified by Nanodrop 1000 spectrophotometer. DNA of all 13 genotypes were diluted to 20 ng/µl with nuclease free water and kept at 4 °C.
RAPD amplification analysis
RAPD amplification was executed according to Williams et al. [20] by decamer random primers (MWG Operon technologies, Germany) (Table 2). RAPD amplification was executed in a 25 µl total volume containing 100 ng DNA, 2.5 µl 10× DreamTaq Green PCR buffer, 40 µM MgCl2, 5 µM dNTPs mix, 1.5U DNA polymerase (KapaBiosystems) and 1 µl of 10 pmol of primer and 15.7 µl distilled water. Amplification was automated using PCR machine (Applied Biosystems, USA). The PCR cycles comprised an initial 94 °C for 5 min for denaturation followed by 42 cycles of denaturation at 94 °C for 1 min, primer annealing at 38 °C for 1 min, extension at 72 °C for 1 min and finally 72 °C for 7 min. Agarose gel (1.5 %) was prepared to separate the amplified product. Gel documentation system (Alpha Innotech, USA) was used for visualization of amplified DNA fragments. Each experiment was repeated three times with each primer to test the reproducibility of RAPD primer.
ISSR amplification analysis
ISSR amplification was performed according to Sarla et al. [21]. ISSR amplification was executed in a 25 µl reaction mixture volume having 100 ng DNA, 2.5 µl 10× DreamTaq Green PCR buffer, 40 µM dNTPs mix, 1.5 U DNA polymerase (KapaBiosystems) and 1 µl of 10 pmol of primer (Table 3) and 15.7 µl distilled water. The amplification reaction involved an initial 94 °C for 5 min for denaturation followed by 35 cycles of 1 min at 94 °C (denaturation), 45 s at a specific annealing temperature 55 °C, and 1 min at 72 °C (extension) and finally 72 °C for 7 min. Agarose gel (1.5 %) was prepared to separate the amplified product. Gel documentation system (Alpha Innotech, USA) was used for visualization of amplified DNA fragments. Each experiment was repeated three times with each primer to test the reproducibility of ISSR primer.
Statistical data analysis
PCR amplification product of RAPD and ISSR markers across the 13 genotypes were scored as presence (1) or absence (0) of band and formed the binary matrix. The faded or unclear fragments were not counted. The percentage of polymorphism was calculated by dividing the polymorphic fragments by the total number of fragments multiplied with 100 by using binary matrix. The polymorphism information content (PIC) was calculated by the formula: PIC = 2π (1 − π) where, π is the frequency of presence of polymorphic fragments in different primers [22]. Similarity matrices were constructed according to Jaccard’s coefficient of similarity by using NTSYS-pc [23]. The Mantel matrix correspondence test was used to estimate the correlation between matrices produced from two marker types [24]. The unweighted pair group method with arithmetic average (UPGMA) was used in construction of dendrogram by using NTSYS-pc [25]. Eigen value and Eigen vectors from a similarity matrix was used in calculation of principle component analysis (PCA) by using NTSYS-pc software.
Results
RAPD amplification analysis
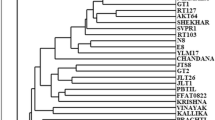
Initially total 120 RAPD primers were screened, out of which 30 primers responded with six or more reproducible bands, were included in the present study. Total 645 DNA bands were produced in all 13 genotypes by PCR amplification of DNA using 30 primers. Amplified fragments varied from six (OPA-02, OPC-02, OPD-18, OPD-M4, OPN-01, OPO-15) to twenty-nine (OPM-11) in number, with the size range of 285-2168 bp. Out of 645 amplified bands, 623 bands were found polymorphic, with an average of 20.76 polymorphic bands per primer. The polymorphism percentage varied from 80.95 (OPC-04) to 100 (OPA-02, OPA-04, OPA-07, OPA-10, OPA-20, OPC-08, OPD-13, OPD-18, OPM-05, OPM-12, OPN-02, OPN-04, OPN-10, OPN-11, OPN-13, OPO-15, OPO-20), with an average of 96.52 % polymorphism. PIC value ranged from 0.85(OPN-15) to 0.96 (OPM-11). The amount of polymorphism detected among all the Indian bamboo genotypes as observed by RAPD primer OPC-04 (Fig. 1a) and OPM-02 (Fig. 1b) are shown. A dendrogram based on UPGMA analysis grouped thirteen bamboo genotypes in the three main clusters (Fig. 2a), with Jaccard’s similarity coefficient of 0.19–0.83 (Table 4). Clusters I comprise GAU-1, GFC-2, and GSF-5, cluster II comprise GKU-3, GKU-4,GVP-6, and OCT-9, and cluster III comprise YKE-11,GNU-7, OVP-8,YNU-10,YVI-12, and YVP-13. The arithmetic mean heterozygosity (Hav) of a marker system indicates its ability to detect heterozygosity in the germplasm and marker index (MI) the parameter specifically used for comparing the utility of two or more marker systems (24). The average similarity coefficient among all genotypes, Hav and marker index (MI) were 0.33, 0.92 and 11.79, respectively. The cophenetic correlation coefficient was positively correlated to the Mantel test statistics in the present study. Also the Mantel Test statistics Z was normalized and extent of goodness of fit for cluster analysis, (matrix correlation r = 0.94) as categorized by Rholf [25] was found to fall under the category ‘good’. Relationship among the 13 genotypes was also detected by PCA based on RAPD data. Genotypes grouped within the same cluster in the dendrogram were occupying the same positions in the three dimensional scaling also (Fig. 3a).
Amplification pattern of 13 Indian genotypes of bamboo obtained with a RAPD primer OPC -04, b RAPD primer OPM- 02, c ISSR primer ISD- 16 and d ISSR primer ISD- 28. Lane M 1-kb ladder and lanes 1–13 represent Indian bamboo genotypes as enumerated in Table 1
ISSR amplification analysis
Initially total 63 ISSR primers were screened, out of which 12 primers responded with more than six scorable bands and gave fingerprints with good resolutions and reproducibility. On the basis of sequence repeat motifs, ISSR primers amplified different numbers of DNA fragments (Fig. 1c, d). A total 12 ISSR primers formed 246 bands across 13 genotypes, of which 241 band were polymorphic, accounting polymorphism of 97.96 %.The number of amplified bands varied from six (ISD4, ISD9, ISD13, ISD20, ISD28, ISD 32, ISD40, UBC 824, UBC 834, UBC 855) to 24 (ISD4) and varied in size from 224 to 2171 bp. The average polymorphic bands and number of bands per primers were observed 20.08 and 20.5 respectively. The average 98.20 % polymorphism was observed. PIC value ranged from 0.84 (UBC 824) to 0.95 (ISD 16) (Table 3). The level of polymorphism detected across all the bamboo genotypes as observed by ISSR primer ISD-16 (Fig. 1c) and ISD-28 (Fig. 1d) are shown. The clustering pattern was similar to RAPD except OCT-9. In RAPD, OCT-9 belong to cluster II whereas, in ISSR, it belong to cluster III (Fig. 2b).The average similarity coefficient among all genotypes, Hav, and MI were 0.30, 0.91 and 7.47, respectively. Also the Mantel test statistic Z was normalized and extent of Goodness of fit for cluster analysis (matrix correlation r = 0.96) as categorized by Rholf [25] was found to fall under the category ‘good’. Relationship among all the 13 genotypes was also resolved by PCA based on ISSR data. The three dimensional scaling of genotypes was in accordance with the clustering pattern found in dendrogram (Fig. 3b) and Jaccard’s similarity coefficient (Table 5).
Pooled RAPD and ISSR analysis
For UPGMA cluster analysis, both RAPD and ISSR data were pooled. From the cluster analysis of pooled ISSR and RAPD data analysis, UPGMA dendrogram was obtained having Jaccard’s similarity coefficient of 0.16–0.84 (Table 6). Clustering pattern of dendrogram generated by using the pooled molecular data of 30 RAPD and 12 ISSR loci was similar to RAPD (Fig. 2c). It was also similar to ISSR except OCT-9. In ISSR, OCT -9 belong to cluster III whereas, in pooled RAPD and ISSR, it belong to cluster II. The average similarity coefficient among all genotypes, Hav and MI were 0.32, 0.92 and 15.64, respectively. Relationship among all the 13 genotypes was also shown by PCA based on pooled RAPD and ISSR data. The genotypes occupied the same position in three dimensional plot as those found in case of clustering pattern of dendrogram (Fig. 3c).
Discussion
Identification of genetic relationships or genetic divergence in bamboo is very tough due to the absence of phenotypic variances and unpredictable flowering. Reliable identification of genotypes or taxa is essential for propagators and consumers, and breeders to confirm protection of intellectual property right. The best conventional technique for detecting species or genotypes by phenotypic features is now swapped by DNA profiling mostly because of numerous limitations of phenotypic data. Both RAPD and ISSR markers are used in study of diverse population [26–28]. In the present study, total 846 polymorphic amplified bands were produced from 30 RAPD and 12 ISSR primers that clearly differentiated 13 bamboo genotypes into two major clusters. More than 60 % polymorphism was observed using both ISSR and RAPD markers. Marker index for RAPD (11.79) is higher than the ISSR marker (7.47) which indicates more specific amplification of the DNA in case of RAPD than ISSR. Matrices were highly correlated in the category ‘good’ for both the markers. Our results detected the occurrence of extensive genetic diversity across 13 genotypes of bamboo. Clustering pattern derived using both the markers were found more or less similar when compared to the pooled RAPD and ISSR dendrogram. Results clearly illustrated that both DNA and ISSR markers were similarly efficient for phylogenetic and diversity studies. However, it was also be concluded that large number of factors like polymorphism percentage, marker index, PIC, and heterozygosity are need to be calculated to judge the best marker. Also, based on RAPD and ISSR data, both phylogenetic and PCA analysis differentiate that all yellow coloured bamboo genotypes were grouped in the same cluster in both the case of RAPD and ISSR. OCT-9 which is an ornamental bamboo with short internodes was clustered distantly and differentially in both the case RAPD and ISSR. A wide range of variation found within Dendrocalamus species clustering D. brandisii very closely within the bambusa species while D. giganteus cluster distantly from the rest of the Bambusa species by using RAPD markers in the study [29]. However, on the basis of similarity matrix, percent polymorphism (RAPD = 96.52; ISSR = 98.20), and PIC values (RAPD = 0.93; ISSR = 0.91), RAPD marker was slightly more revealing than ISSR in the evaluation of genetic diversity in bamboo. Gajera et al. [15] has also observed similar results in castor. This may be due to the fact that different parts of the genome are targeted by different two markers systems. Some investigators have considered that RAPD markers characterize non-coding portions of DNA and to be selectively neutral [30, 31], and some researchers have revealed that RAPD markers are disseminated throughout the genome and linked with functionally main loci [32]. Nevertheless, there is less information to specify that ISSR markers are functionally significant [21]. Present investigation indicates the existence of wide genetic diversity between different genotypes of bamboo. Both DNA based molecular markers (RAPD and ISSR) are suitable in the evaluation of bamboo genetic diversity, the revealing of identical sample in population, and the detection of a core population to improve the efficacy of genotype management for use in bamboo breeding and conservation. The genotype specific diagnostic bands had also been identified using RAPD and ISSR markers which could be transformed into SCAR markers for discriminations of these genotypes after further validation of those markers by using more numbers of genotypes.
References
Wang D, Shen SJ (1987) Bamboos of China. Timber Press, Portland
Thomas TA, Arora RK, Singh R (1988) Genetic wealth of bamboos in India and their conservation strategies. Bamboos current research. In: Proceedings of international bamboo workshop, Cochin, 32 Nov 1988, pp 14–18
Loh JP, Kiew R, Set O, Gan LH, Gan YY (2000) A study of genetic variation and relationship within the bamboo subtribe Bambusinae using amplified fragment length polymorphism. Ann Bot 85:607–612
Bahadur KN (1979) Taxonomy of bamboos. Ind J For 2:222–241
Soderstorm TR, Calderon CE (1979) A commentary on bamboos (Poaceae: bambusoideae). Biotropica 11:161–172
Yeasmin L, Ali MN, Gantait S, Chakraborty S (2014) Bamboo: an overview on its genetic diversity and characterization. 3 Biotech. doi:10.1007/s13205-014-0201-5
Stapleton CM, Rao VR (1995) Progress and prospects in genetic diversity studies on bamboo and its conservation. Bamboo, people and the environment. In: Proceedings of Vth international bamboo workshop and the IVth international bamboo congress, Ubud, Bali, 19–22 June 1995
Nayak S, Rout GR, Das P (2003) Evaluation of genetic variability in bamboo using RAPD markers. Plant Soil Environ 49(1):24–28
Goyal AK, Ghosh PK, Dubey PK, Sen A (2012) Inventorying bamboo biodiversity of North Bengal: a case study. Int J Fund Appl Sci 1:5–8
Welsh J, McClelland M (1990) Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res 18:7213–7218
Gwanama C, Labuschagne MT, Botha AM (2000) Analysis of genetic variation in Cucurbita moschata by random amplified polymorphic DNA (RAPD) markers. Euphytica 113:19–24
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238
Kapteyn J, Simon JE (2002) The use of RAPDs for assessment of identity, diversity and quality of Echinacea. In: Janick J, Whipkey A (eds) Trends in new crops and new uses. ASHS Press, Alexandria, pp 509–513
Arif M, Zaidi NW, Singh YP, Haq QMR, Singh US (2009) A comparative analysis of ISSR and RAPD markers for study of genetic diversity in Shisham (Dalbergia sissoo). Plant MolBiol Rep 27:488–495
Gajera BB, Kumar N, Singh AS, Punvar BS, Ravikiran R, Subhash N, Jadeja GC (2010) Assessment of genetic diversity in castor (Ricinus communis L.) using RAPD and ISSR markers. Indus Crops Prod 32:491–498
Kong Q, Li X, Xiang C, Wang H, Song J, Zhi H (2011) Genetic diversity of Radish (Raphanus sativus L.) germplasm resources revealed by AFLP and RAPD markers. Plant Mol Biol Rep 29:217–223
Gonzalez A, Coulson A, Brettell R (2002) Development of DNA markers (ISSRs) in mango. Acta Hortic 575:139–143
Lin XC, Ruan XS, Lou YF, Guo XQ, Fang W (2008) Genetic similarity among cultivars of Phyllostachys pubescens. Plant Syst Evol 277:67–73
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Williams JGK, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphism amplified by arbitrary primers are useful as genetic markers. Nucl Acid Res 18:6531–6535
Sarla N, Bobba S, Siddiq EA (2003) ISSR and SSR markers based on AG and GA repeats delineate geographically diverse Oryza nivara accessions and reveal rare alleles. CurrSci 84(5):683–690
Bhat KV (2002) Molecular data analysis. In: Proceedings of the short-term training course on molecular marker application in plant breeding, ICAR, New Delhi, 26 Sep–5 October 2002
Rohlf FJ (1993) NT-SYS-pc: Numerical taxonomy and multivariate analysis system, version 2.11W. Exteer software, Setauket
Mantel N (1967) The detection of disease clustering and generalized regression approach. Cancer Res 27:209–220
Sneath PHA, Sokal RR (1973) Numerical taxonomy. Freeman Press, San Francisco
Li A, Ge S (2001) Genetic variation and clonal diversity of Psammochloa villosa (Poaceae) detected by ISSR markers. Ann Bot 87:585–590
Esselman EJ, Li JQ, Crawford D, Winduss JL, Wolfe AD (1999) Clonal diversity in the rare Calamagrostis porteri ssp. Insperata (Poaceae): comparative results for allozymes and random amplified polymorphic DNA (RAPD) and inter-simple sequence repeat (ISSR) markers. Mol Ecol 8:443–451
Parsons BJ, Newbury HJ, Jackson MT, Ford-Lloyd BV (1997) Contrasting genetic diversity relationships are revealed in rice (Oryza sativa L.) using different marker types. Mol Breed 3:115–125
Eevera T, Rajandran K, Saradha S, Lashmi A (2008) Ananlysis of genetic variation in selected bamboo species using RAPD. Tree For Sci Biotechnol 2(1):54–56
Bachmann K (1997) Nuclear DNA markers in plant biosystematics research. Opera Bot 132:137–148
Landergott U, Holderegger R, Kozlowski G, Schneller JJ (2001) Historical bottlenecks decrease genetic diversity in natural populations of Dryopteris cristata. Heredity 87:344–355
Penner GA (1996) RAPD analysis of plant genomes. In: Jauhar PP (ed) Methods of genome analysis in plants. CRC, Boca Raton, pp 251–268
Acknowledgments
The authors are thankful to division of Food Corporation of India, Anand, Gujarat, S. P. University, V.V. Nagar, Gujarat, Anand Agricultural University, Anand, Gujarat, Farmers of Kutch region, Gujarat and Rajiv Gandhi Centre for Biotechnology, Thiruvananthapuram, Kerala for providing the different genotypes of bamboo.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Desai, P., Gajera, B., Mankad, M. et al. Comparative assessment of genetic diversity among Indian bamboo genotypes using RAPD and ISSR markers. Mol Biol Rep 42, 1265–1273 (2015). https://doi.org/10.1007/s11033-015-3867-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-015-3867-9