Abstract
Strong anthropogenic impact has caused 28 of the currently recognized 55 species of deer (Cervidae) to be listed on the IUCN Red List. Particular threats to vulnerable species include habitat deterioration and hybridization with alien, introduced species. The scarcity of many species has severely hampered genetic analyses of their populations, including the detection of loci for cross-species amplification. Because deer antlers are shed and re-grown annually, antlers offer the possibility for non-invasive genetic sampling of large individual numbers, and may provide material for reference genotyping from historical samples stored in zoos, museums and trophy collections of rare and extinct species/populations. In this paper, we report cross-species amplification of 19 nuclear microsatellite loci and the amplification of 16S mtDNA for barcoding from nearly a third of all deer species worldwide based on high quality DNA extracted from antler bone up to 40 years old. Phylogenetic analysis based on mtDNA of seventeen species and five subspecies corroborate previously published phylogenetic data, thus confirming the specific resolution of the DNA extraction methodology.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The deer family (Cervidae) is distributed throughout the northern hemisphere as well as in South America and southeast Asia [1]. With 55 species recognized by the International Union for the Conservation of Nature (IUCN), the family constitutes the second most species-rich family of artiodactyls after the Bovidae [2]. Despite their wide distribution, half of all deer species are today red listed by the IUCN as vulnerable, endangered, critically endangered or extinct in the wild. Overexploitation of natural populations (illegal hunting, etc.) and habitat loss have led to the severe population reductions. Furthermore, deer products, especially antlers and antler powder, are widely employed in animal-based traditional Chinese medicines. Deer antlers are used to treat ailments ranging from anemia and lumbago to impotence [3]. Consequently, antlers in fresh velvet can cost up to $1,000 per kg.
The economic significance of deer as managed game animals, for trophy hunting and commercial meat and antler production have led to numerous translocations of deer species worldwide [4]. Translocations may have severe consequences for the conservation of indigenous species [4] due to interspecific competition and/or the introduction of new diseases and parasites [5, 6], both of which may lead to a reduction in the density of native species. Translocated individuals might also hybridize with native populations of the same or closely related species, consequently impacting the native gene pool of each species. For example, introduced Sika deer (Cervus nippon) readily interbreed with native red deer (Cervus elaphus) in parts of Great Britain to such an extent that no native red deer without some degree of Sika deer ancestry are currently found in these areas [7, 8].
Cervidae are united by a series of synapomorphies, including the possession of antlers in males [9]. Antlers are made of a deciduous bony core covered by velvet skin. Antlers are shed annually and regenerate in the following 5 months from the permanent pedicles [10]. The annual shedding and renewing makes antler an ideal material for non-invasive sampling of deer species. Non-invasive sampling allows for investigations into the biology of elusive, rare and/or endangered species [11] without harming and disturbing or even having to observe individuals. It is therefore of special importance in conservation biology [12]. In a previous paper, Hoffmann and Griebeler [13] established an improved method to isolate and analyze high quality DNA from antlers of red deer and they successfully used the DNA for microsatellite genotyping. In the current paper, we show the applicability of this method for the extraction of high quality DNA for microsatellite and mtDNA genotyping of antlers from about a third of the world’s deer species, including species extinct in the wild.
Microsatellites are versatile tools to investigate paternity, kinship and population structure in both captive breeding programs and hybridizations in the wild [14, 15]. When testing intraspecific populations, knowledge of species-specific loci is adequate. However, for between-species studies such as hybridization tests or for phylogenetic inference, cross-species amplifying loci are required [16]. Among the many studies using microsatellites to study various species of deer, there are few studies that use the same sets of primers [17]. We therefore used the DNA from the studied deer species to establish a consistent microsatellite marker system with cross-species amplification tests with 19 microsatellite loci previously used in cattle and red deer [18–21]. In total, 17 deer species and five subspecies were genotyped. For some of these species (Cervus albirostris, Rusa unicolor, Dama mesopotamica) and subspecies only a small number of polymorphic loci have been described, until now [17].
We further amplified 550 bp of the mitochondrial 16S rDNA gene to verify the DNA-extraction method’s ability to produce high quality DNA for barcoding (species determination) and phylogeny [22]. Barcoding entails the screening of one mitochondrial reference gene to assign unknown individuals to a species [23]. DNA barcoding, with its unique reproducibility, sequence versatility and comparability among different species, provides a powerful approach for species authentication [24] and enables species determination of unknown samples from various sources such as meat, antlers and trophies, and from the powder of antlers. Our objectives in this study were to characterize the mtDNA 16S rDNA and microsatellite DNA variation from non-invasive tissue sampling (antler) in 17 deer taxa for use in species identification, population genetics and phylogenetic analyses.
Materials and methods
Sampled animals
We conducted cross-amplification tests with 19 microsatellite loci and the mitochondrial 16SrDNA for 42 individuals, comprising 17 species and five subspecies with DNA extracted from antlers. Because the subjective nature of subspecies designations [25] we defined subspecies as geographic populations with a largely independent phylogeny [26]. Most of the antlers originate from the Tierpark Berlin (TB). The origin of all samples and the species’ status on the IUCN Red List are given in Table 1. Although there is no Red List status available for subspecies, all subspecies (except for Cervus canadensis sibiricus) in this study are endangered; for example, C. nippon pseudaxis is extinct in the wild. The threatened status of some of the researched species limited the number of individuals available for our analysis. The number of analysed individuals per species/subspecies was between one and three (Table 1).
DNA extraction from antlers
DNA was extracted from a small borehole drilled on the backside of antlers following the method presented in Hoffmann and Griebeler [13]. The antler chips were pulverized in a Retsch-mill at a frequency of 30 Hz for 120 s. The DNA was therefore extracted from powdered antler samples, similar to that used in traditional Chinese medicine. The DNA content of every sample was measured with a NanoDrop1000 (Peqlab, Erlangen, Germany). Before each PCR, every sample was diluted to a final DNA concentration of 50–100 µg/ml (measured with NanoDrop 1000).
Microsatellite primers, PCR reactions and microsatellite genotyping
For the amplification of the 19 microsatellites we applied two different PCR-based methods. For eleven fluorescently labelled primers, we used the Qiagen Multiplex Kit (Qiagen, Basel, Switzerland) (Haut 14, ILSTS06, BM1818, CSSM14, CSPS115, CSSM16, MM12, Inra35, Cer14, CSSM22 and CSSM19) (all used primers and primer-sequences are listed in Supplementary Table S3). The reaction volume for each sample consisted of 5 µl Qiagen Multiplex PCR Mastermix, containing dNTPs, QiagenHotStarTaq DNA Polymerase and 6 mM MgCl2, 1 µl Primer Mix (2 µM each Primer), 2.5 µl RNase-free water and 1.5 µl DNA (final DNA concentration of 50–100 µg/ml) resulting in a final reaction volume of 10 µl for each sample. PCRs were performed on a Perkin Elmer GeneAmp 9600 thermocycler using the recommended multiplex-PCR protocol (Qiagen) with the following cycling conditions: 15 min at 95 °C for initial denaturation; then 40 cycles of 40 s at 94 °C, 90 s at 57 °C and 90 s at 72 °C, followed by 30 min at 60 °C and a final hold at 4 °C.
The remaining eight microsatellite primers (NVHRT48, RT1, RT6, IOBT965, BM4107, NVHRT16, Ca38, Ca60) were labelled during the PCR reaction following the method in [27]). Each reaction consisted of 7.7 µl H2O with Ready to go PCR Beads (GE Healthcare), ~0,035 µl Forward Primer (100 pM), ~0.14 µl Reward Primer (100 pM), ~0.14 µl M13 Tail (HEX, FAM, NED) and 1 µl DNA resulting in a final reaction volume of 9 µl for each sample. To detect the best annealing temperature we ran gradient PCRs for each primer on a TGradient Biometra Thermocycler (Biometra). Optimal annealing temperatures were: NVHRT48 (53 °C), RT1 (51 °C), RT6 (51 °C), IOBT965 (54 °C), BM4107 (55 °C), NVHRT16 (53 °C), Ca38 and Ca60 (no amplification, see Results). Subsequent PCRs were performed on a Perkin Elmer GeneAmp 9600 thermocycler using a PCR protocol with the following cycling conditions: 5 min at 94 °C for initial denaturation; then 30 cycles of 30 s at 94 °C, 45 s at 51–55 °C and 45 s at 72 °C; then 8 cycles of 30 s at 94 °C, 45 s at 53 °C and 45 s at 72 °C followed by 10 min at 72 °C and a final hold at 4 °C.
Purified microsatellite PCR fragments were run on a 3130xl capillary sequencer (Applied Biosystems) using a GS500 ROX size standard. The genotypes were analysed with GENEMAPPER, version 4.0 (Applied Biosystems). To avoid false allele scoring, we only scored alleles with markedly high fluorescent values and a distinctive, characteristic form at each locus. Samples with ambiguous peaks were assessed as no amplification success. To assess quality and reproducibility of genotypes, we re-extracted DNA and genotyped more than a fourth (n = 11) of all individuals twice (from seven different species). These eleven individuals were amplified across ten loci to calculate the amplification success rate, false allele rate and allelic dropout rate between replications. We did not test for null allele frequencies due to the low number of individuals per species (Table 1).
mtDNA primers, PCR reactions and 16S rRNA genotyping
We amplified the mitochondrial gene 16S rRNA (550 bp, positions 2306–2856 in Bos taurus) using universal primer pairs (fwd 5-CGC CTG TTT ATC AAA AAC AT-3 and rev 5-CTC CGG TTT GAA CTC AGA TC-3) [28]. Each 25 µl PCR reaction consisted of 21 µL of H2O, 1 µL of each respective primer (100 pM) and 2 µL of DNA extract in a 0.5 mL PCR tube with illustra™ puReTaq Ready-To-Go PCR Beads (GE Healthcare). The final volume of 25 µL contained 200 µM of each dNTP, 10 mM Tris–HCL (pH 9.0 at room temperature), 50 mM KCl, 1.5 mM MgCl2, approximately 2.5 units of puReTaq DNA polymerase, reaction buffer and the stabilizers BSA, dATP, dCTP, dGTP and dTTP. Thermocycling included 94 °C for 1 min, 30 cycles of 94 °C for 30 s, 49 °C for 30 s and extension at 72 °C for 45 s. The reaction was stored at 4 °C after a final extension at 72 °C for 10 min. Reactions were performed on a TGradient Biometra Thermocycler (Biometra). PCR products were separated using gel electrophoresis (1 % Agarose); fragments were controlled for expected size and band quality. The amplified PCR fragments were purified using the Roche High Pure PCR Product Purification Kit (Roche Diagnostics).
The primary aim of the 16S sequence analysis was to demonstrate the versatility of the DNA extraction method. We show that the extracted DNA can be used to produce high quality DNA sequences for barcoding and phylogeny. We amplified a 550 bp fragment of the mitochondrial 16S rDNA for all species and subspecies (primer and primer-sequences listed in Supplementary Table S3; accession numbers in Supplementary Table S6). For effective DNA barcoding, sequence variation must be high enough among species to be discriminatory, but low enough within a species so that a clear threshold between intra- and inter-specific genetic variations can be defined [24]. Due to the low number of individuals per species analyzed in this study (Table 1), we blasted the 16S mtDNA sequence for each species in GenBank to evaluate intraspecific sequence variation. We found between two and eleven additional sequences per species (Supplementary Table S1; accession numbers in Supplementary Table S6). 16S sequence is frequently used for various species for phylogenetic studies and therefore applicable for barcoding because of the high numbers of comparable sequences in molecular databases.
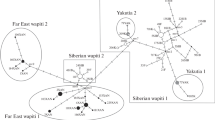
We established a phylogenetic tree based on haplotype sequences and compared the phylogeny to previously published ones [1]. Phylogenetic inference based on the 550 bp 16S rDNA mtDNA fragment was done with Neighbor Joining and Maximum Parsimony using the Tamura 3-Parameter (α = 0.15) evolution model, which was the best model detected by MEGA version 5 [29]. Bos taurus (Bovidae) was used as an outgroup (Accession number: HQ184044.1). Each phylogenetic inference was evaluated with 1,000 bootstraps. Only bootstrap scores higher than 70 % were considered as significant support of branching.
Results
DNA extraction
DNA was successfully extracted from antlers in moderate to high concentrations from all individuals. DNA concentrations ranged between 31.06 and 1,046.23 µg ml−1 (mean: 227.92 µg ml−1, standard deviation: 275.93 (Supplementary Table S2)).
Cross-species amplification of microsatellite loci
Cross-species amplification of microsatellite loci was tested with 19 loci (primer pairs) for all species and subspecies. Of particular interest were species listed in the IUCN Red List and subspecies in danger of extinction, but also species for which only a few microsatellites have been described (Cervus albirostris, Rusa unicolor, Dama mesopotamica). Table 2 summarizes amplification success, and the range and number of alleles found for these specific species. A complete list of amplification success, range and number of alleles for all other species/subspecies is presented in Supplementary Table S4, S7. The tested primers, which were developed for cattle [18–21], showed a high amplification success in all species and subspecies. For each of the investigated species/subspecies, 9–16 primer pairs amplified fragments (Supplementary Table S4, S7). The loci RT1 and RT6 failed to amplify in all species. The number of polymorphic loci ranged from two (C. nippon mantschuricus) to 12 (C. albirostris, C. elaphus). In total, nine primer-pairs amplified fragments in all examined species (CER14, NVHRT48, RT1, MM12, INRA35, CSSM22, CSSM19, BM1818, CSPS115).
The test for reproducibility of microsatellite genotyping with 11 individuals at ten loci resulted in a false allele rate between replicates of 2.2 % and in the dropout or no amplification of one allele in 1.4 % of all replicates. Genotypes were analyzable and amplification success in the first replication was at 96.4 % across all of the samples.
16S rDNA
The mtDNA 16S rRNA fragment (550 bp) was successfully amplified in all individuals from DNA extracted from antlers (Supplementary Table S5). To compare specific/subspecific sequence diversity for barcoding, we searched GenBank (search date 29.05.2014) and compared the nucleotide diversity for each species with our samples (number of samples per species and intraspecific variation in Supplementary Table S1). Sequence diversities here mean the % of the nucleotides that differ between two sequences. The mean intraspecific haplotype variation across all species, including blasted sequences from GenBank, was 0.6 % across the 550 nucleotides. Between species, the genetic variation ranged from 0 % (Odocoileus hemionus, O. virginianus) to 6.8 %. Except for O. hemionus and O. virginianus, no species shared the same haplotype sequence. To further evaluate the reproducibility and phylogenetic accuracy of the 16S sequence data, we calculated phylogenetic trees applying the Neighbor Joining and Maximum Parsimony method (Fig. 1). The phylogenetic trees are concordant with known phylogenies of Cervidae, though the strict monophyly of Plesiometacarpalia and Metacarpalia is not shown.
Discussion
We successfully extracted nuclear and mitochondrial DNA of high quality from antlers of nearly half of the world’s cervid species and we used the DNA to establish a microsatellite marker system and 16S mtDNA barcoding inventory. While the extraction method was previously limited to the successful amplification of red deer nuclear DNA [13], here we provide proof of the method’s applicability to study a wide range of deer species worldwide for conservation and other scientific interests. The astoundingly high concentration and quality of the DNA of all deer species studied can be explained by the almost perfect storage of the antlers as trophies. The fact that antler tissue is desiccated before antlers are shed [13] protects the DNA from breakdown into cellular compartments and degradation by catabolic enzymes, e.g. lysosomal nucleases [30]. Easy sampling and the small amounts of bone tissue (0.2 g) needed for analyses makes our method readily applicable for fast and efficient sampling and genotyping of numerous individuals, deer products, antlers, trophies and powder, as well as hunted or poached individuals. No expensive or special equipment is needed for sampling. In addition, the small borehole drilled on the backside of trophies is not visible, which is important when sampling from rare or valuable trophies.
Although we cannot make general statements about the extraction method’s applicability for non-antler bone, we have successfully extracted DNA from jaw (80 year old) and femur bone stored in museums (GSH, unpublished data).
Antlers in this study were between two and 40 years old. For this reason, we cannot attest to DNA preservation >4 decades in each species, as evidenced in red deer antlers up to 200 years old [13]. Nevertheless, the high structural similarly of cervid antlers indicates that DNA preservation will be equally good in all other species. The high quality of DNA in antlers, preserved for at least 200 years, offers the opportunity for non-invasive genotyping of rare species over time, reconstructing histories of extinct populations and translocations of deer worldwide, without observing individuals [11, 12]. Non-invasive techniques are of great conservation concern and are now routinely used; for instance, feces is used to monitor brown bears in North America [31, 32]. For cervid populations in captive breeding programmes with or without complete breeding books, the determination of kinship from old shed antlers or trophies is possible.
In our effort to validate the quality and applicability of DNA extracted from antlers as a reliable tool for non-invasive genotyping, we established a microsatellite marker system for different cervid species/subspecies. The 19 microsatellite loci were originally developed for cattle and some have already been used individually in cervids. For Cervus albirostris, Rusa unicolor and Dama mesopotamica only a few polymorphic microsatellites were described until now [17]. We found between 9 and 16 primer-pairs that amplified between 2 and 12 polymorphic loci for each of these species. While the number of polymorphic loci was not formerly sufficient for population genetic studies, we could increase the number of applicable polymorphic primers for these threatened species. For the remaining species, for which microsatellite loci are already known, our new marker system identified loci/primers for inter- and intraspecific studies, including conservation and forensic studies [33, 34]. We are well aware that testing 1–3 individuals per species does not capture the genetic diversity of a species or the diagnostic value of specific alleles among closely related species. The low sample size underestimates the number of polymorphic loci, which might also be due to inbreeding in captivity. We therefore can expect that the number of polymorphic loci per species is a lower bound. With heterologous microsatellites, issues with amplification and consistency of scoring might not be apparent until many individuals are scored. Due to low sample size we were also unable to address potential null alleles. There was indeed a high ratio of homozygotes (and monomorphic loci) in the sample. However, based on low sample sizes it is impossible to rule out whether there are technical reasons for this phenomenon (false alleles, bovine origin of primers) or biological/genetic reasons like null alleles and inbreeding, or if some of the loci are less polymorphic in general. We further assessed the non-invasive genotyping potential of the sampling method based on its ability to produce high quality sequence data for, e.g., barcoding and phylogeny. DNA barcoding must have a low threshold and high efficiency to make it a sensitive method for the identification of different species [35] and animal derived compounds. mtDNA provides this opportunity and was therefore used for cervid species identification [22]. Accurate species identification requires a comprehensive comparative molecular database for the assignment of an unknown individual [35]. In our study, amplification of the 16S gene produced unambiguous sequences with low mean intra-specific sequence diversities (<0.1 %; under 0.8 % of the nucleotides differ between two sequences when including sequences found in GenBank), whereas inter-specific diversity was much higher (mean 3.3 %). In accordance with another study [22], only the two common species O. hemionus and O. virginianus could not be differentiated because they had identical haplotypes.
The phylogenetic tree calculated with 16S (Fig. 1) confirmed that (1) individuals of the same species cluster together, while individuals of different species are discriminated and (2) that cervid species in this study are mostly divided in two major clades, Plesiometacarpalia and Metacarpalia, as found in previous studies [22, 30]. The fact that the strict monophyletic origin of these two subfamilies is not shown (Odocoileus and Rangifer occur in the large clade with Metacarpalia in the Maximum Parsimony Tree), as also seen in the paraphyletic mtDNA phylogeny within Odocoileus [36], reflects the limitations of single gene trees for phylogeny inference. Nevertheless for our limited analysis of only 550 bp, results are good and we thus conclude that the 16S fragment is efficient in identifying (barcoding) specific species of deer because of high inter-specific and low intra-specific variance; the latter is also evidenced in a larger sample of red deer individuals from different geographical regions in central Europe (Germany and Austria) (GS Hoffmann, unpublished results).
In summary, the extraction method in combination with the different markers used offers a versatile tool for not only species determination, forensic analyses and assessing trafficking of rare species but also is for non-invasive population genetic and conservation studies. A further advantage of the method is that is possible to discriminate species from antler powders, such which are normally used in traditional Chinese medicine. All DNA samples used in this study were extracted from powdered antler samples (see Materials and Methods).
References
Gilbert C, Ropiquet A, Hassanin A (2006) Mitochondrial and nuclear phylogenies of Cervidae (Mammalia, Ruminantia): systematics, morphology, and biogeography. Mol Phylogenet Evol 40(1):101–117. doi:10.1016/j.ympev.2006.02.017
Grubb P (1993) Order artiodactyla. In: Wemmer CM (ed) Mammal species of the world: a taxonomic and geographic reference, 2nd edn. JHU Press, Maryland, pp 377–414
Mainka SA, Mills JA (1995) Wildlife and traditional Chinese medicine—supply and demand for wildlife species. J Zoo Wildl Med 26(2):193–200
Galindoleal C, Weber M (1994) Translocation of deer subspecies—reproductive implications. Wildl Soc Bull 22(1):117–120
Geist V (1988) How markets in wildlife meat and parts and the sale of hunting privileges jeopardize wildlife conservation. Conserv Biol 2(1):15–26. doi:10.1111/j.1523-1739.1988.tb00331.x
Geist V (1992) Deer ranching for products and paid hunting: threat to conservation and biodiversity by luxury markets. The biology of deer. Springer, New York, pp 554–561
Lower VPW, Gardiner AS (1975) Hybridization between red deer (Cervus-elaphus) and sika deer (Cervus-nippon) with particular reference to stocks in NW England. J Zool 177(4):553–566
Senn HV, Pemberton JM (2009) Variable extent of hybridization between invasive sika (Cervus nippon) and native red deer (C-elaphus) in a small geographical area. Mol Ecol 18(5):862–876. doi:10.1111/j.1365-294X.2008.04051.x
Janis CM, Scott KM (1987) The interrelationships of higher ruminant families with special emphasis on the members of the cervoidea. Am Mus Novit 2893:1–86
Li CY, Suttie JM (2001) Deer antlerogenic periosteum: a piece of postnatally retained embryonic tissue? Anat Embryol 204(5):375–388. doi:10.1007/s004290100204
Piggott MP, Taylor AC (2003) Remote collection of animal DNA and its applications in conservation management and understanding the population biology of rare and cryptic species. Wildl Res 30(1):1–13. doi:10.1071/wr02077
Broquet T, Ménard N, Petit E (2007) Noninvasive population genetics: a review of sample source, diet, fragment length and microsatellite motif effects on amplification success and genotyping error rates. Conserv Genet 8(1):249–260. doi:10.1007/s10592-006-9146-5
Hoffmann GS, Griebeler EM (2013) An improved high yield method to obtain microsatellite genotypes from red deer antlers up to 200 years old. Mol Ecol Resour 13(3):440–446. doi:10.1111/1755-0998.12068
Bruford MW, Wayne RK (1993) Microsatellites and their application to population genetic studies. Curr Opin Genet Dev 3(6):939–943. doi:10.1016/0959-437x(93)90017-j
Bruford MW, Cheesman DJ, Coote T, Green HA, Haines SA, O’Ryan C, Williams TR (1996) Microsatellites and their application to conservation genetics. Molecular genetic approaches in conservation. Oxford University Press, New York, pp 278–297
Goodman SJ, Barton NH, Swanson G, Abernethy K, Pemberton JM (1999) Introgression through rare hybridization: a genetic study of a hybrid zone between red and sika deer (genus Cervus) in Argyll. Scotl Genet 152(1):355–371
Bonnet A, Thevenon S, Maudet F, Maillard JC (2002) Efficiency of semi-automated fluorescent multiplex PCRs with 11 microsatellite markers for genetic studies of deer populations. Anim Genet 33(5):343–350. doi:10.1046/j.1365-2052.2002.00873.x
Kuehn R, Schroeder W, Pirchner F, Rottmann O (2003) Genetic diversity, gene flow and drift in Bavarian red deer populations (Cervus elaphus). Conserv Genet 4(2):157–166. doi:10.1023/a:1023394707884
Poetsch M, Seefeldt S, Maschke M, Lignitz E (2001) Analysis of microsatellite polymorphism in red deer, roe deer, and fallow deer—possible employment in forensic applications. Forensic Sci Int 116(1):1–8. doi:10.1016/s0379-0738(00)00337-6
Goodman SJ, Tamate HB, Wilson R, Nagata J, Tatsuzawa S, Swanson GM, Pemberton JM, McCullough DR (2001) Bottlenecks, drift and differentiation: the population structure and demographic history of sika deer (Cervus nippon) in the Japanese archipelago. Mol Ecol 10(6):1357–1370. doi:10.1046/j.1365-294X.2001.01277.x
Gaur A, Singh A, Arunabala V, Umapathy G, Shailaja K, Singh L (2003) Development and characterization of 10 novel microsatellite markers from Chital deer (Cervus axis) and their cross-amplification in other related species. Mol Ecol Notes 3(4):607–609. doi:10.1046/j.1471-8286.2003.00528.x
Cronin MA, Palmisciano DA, Vyse ER, Cameron DG (1991) Mitochondrial DNA in wildlife forensic science: species identification of tissues. Wildl Soc Bull: 94–105
Hebert PDN, Cywinska A, Ball SL, DeWaard JR (2003) Biological identifications through DNA barcodes. Proc R Soc B Biol Sci 270(1512):313–321. doi:10.1098/rspb.2002.2218
Yan D, Luo JY, Han YM, Peng C, Dong XP, Chen SL, Sun LG, Xiao XH (2013) Forensic DNA barcoding and bio-response studies of animal horn products used in traditional medicine. PLoS One 8(2):e5854. doi:10.1371/journal.pone.0055854
Cronin MA (2006) A proposal to eliminate redundant terminology for intra-species groups. Wildl Soc Bull 34(1):237–241 10.2193/0091-7648(2006) 34[237:aptert]2.0.co;2
Ball RM, Avise JC (1992) Mitochondrial-dna phylogeographic differentiation among avian populations and the evolutionary significance of subspecies. The Auk 109(3):626–636
Schuelke M (2000) An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol 18(2):233–234. doi:10.1038/72708
Guha S, Goyal SP, Kashyap VK (2006) Genomic variation in the mitochondrially encoded cytochrome b (MT-CYB) and 16S rRNA (MT-RNR2) genes: characterization of eight endangered Pecoran species. Anim Genet 37(3):262–265. doi:10.1111/j.1365-2052.2006.01421.x
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739. doi:10.1093/molbev/msr121
Paabo S, Poinar H, Serre D, Jaenicke-Despres V, Hebler J, Rohland N, Kuch M, Krause J, Vigilant L, Hofreiter M (2004) Genetic analyses from ancient DNA. Annu Rev Genet 38:645–679. doi:10.1146/annurev.genet.37.110801.143214
Woods JG, Paetkau D, Lewis D, McLellan BN, Proctor M, Strobeck C (1999) Genetic tagging of free-ranging black and brown bears. Wildl Soc Bull 27(3):616–627
Paetkau D (2003) An empirical exploration of data quality in DNA-based population inventories. Mol Ecol 12(6):1375–1387. doi:10.1046/j.1365-294X.2003.01820.x
Felmer DR, Sagredo DB, Chavez RR, Iraira HS, Folch MC, Parra GL, Catrileo SA, Ortiz LM (2008) Implementation of a molecular system for traceability of beef based on microsatellite markers. Chil J Agric Res 68(4):342–351
Teletchea F, Bernillon J, Duffraisse M, Laudet V, Hanni C (2008) Molecular identification of vertebrate species by oligonucleotide microarray in food and forensic samples. J Appl Ecol 45(3):967–975. doi:10.1111/j.1365-2664.2007.01415.x
Meyer CP, Paulay G (2005) DNA barcoding: error rates based on comprehensive sampling. PLoS Biol 3(12):2229–2238. doi:10.1371/journal.pbio.0030422
Cronin MA, Vyse ER, Cameron DG (1988) Genetic-relationships between mule deer and white-tailed deer in montana. J Wildl Manag 52(2):320–328. doi:10.2307/3801243
Acknowledgments
We are grateful to Christian Kern and the “Tierpark Berlin” for supporting the sampling of specimens. We thank Silke Gabel-Scheurich, Dagmar Klebsch and Ariana Macon for technical assistance. We are also very grateful to Rebecca Nagel for linguistic improvement of the manuscript. This manuscript is part of the Ph.D. thesis of GSH.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hoffmann, G.S., Johannesen, J. & Griebeler, E.M. Species cross-amplification, identification and genetic variation of 17 species of deer (Cervidae) with microsatellite and mitochondrial DNA from antlers. Mol Biol Rep 42, 1059–1067 (2015). https://doi.org/10.1007/s11033-014-3845-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3845-7