Abstract
MicroRNA (miRNA) is a large class of non-coding RNA which usually acts a fine-tuned regulation in repressing gene expression on transcriptional and post-transcriptional level during ontogenetic development, metabolism, and occurrence of disease. Due to the lateness of goat genome investigation, registered goat microRNAs are little known and the function of it is poorly understood. In this study, we identified 5 novel miRNAs express in cashmere goat skin and longissimus dorsi using comparative genomic approach combined with expression profile analysis. Further qPCR and sequencing validation demonstrated that the novel miRNAs we identified expressed in goat skin and longissimus dorsi, three of them with the highest expression in February and two in October, indicating these novel microRNAs were involved in growth and cessation of goat hair production. Subsequently, potential target genes were explored via base-pairing with complementary sequences miRNA/mRNA interaction searching. WDR12 and CSNK1A1 involved in Notch/Wnt signal transduction pathway were finally identified. Collectively, the approach of comparative genomics combining expression profile analysis is a powerful tool to identify novel miRNA in goat and the 5 identified novel miRNAs(chi-miR-2284n, chi-miR-421*, chi-miR-421, chi-miR-1839 and chi-miR-374; Accession number: JQ002550–JQ002554) play a essential role in goat hair production of cashmere goat, both in entering growth and cessation phase. This study is also a meaningful complement to the present goat miRNA database for further understanding function of miRNAs in regulation of goat cashmere production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
MicroRNA (miRNA), first discovered in 1993 by Victor Ambros, Rosalind Lee and Rhonda Feinbaum [1] when investigating the gene lin-14 in Caenorhabditis elegans development, is a large class of small non-coding RNA molecule that plays important role in the process of cell growth and apoptosis, blood cell differentiation, homeobox gene regulation, the polarity of neurons, insulin secretion, brain morphogenesis, heart development and in the latter stages of embryonic development [2] by repressing target gene expression in a fine-tuned manner on transcriptional and post-transcriptional level [3, 4]. Typical mature miRNA is usually 22 nt in length and processed by endogenous dicer enzyme by cleaving hairpin and double strands immaturely pre-miRNA transcripts. As a part of RNA-induced silencing complex (RISC), miRNA anchor to 3′UTR of specific target genes mRNA via base-pairing with complementary sequences within mRNA molecules, usually resulting in gene silencing via translational repression or target degradation [4–6].
As much as 40 % of miRNAs genes located in the intron of protein and non-protein coding genes or even in exon of long nonprotein-coding transcripts [7], with a significant feature of evolutional conservation across many species, it facilitate us the possibilities to predict novel miRNAs by comparative genomics approach base on conservation property.
Cashmere goat is a breed of goat with great value of producing large number high quality cashmere generated in specific condition by natural selection and artificially reproduces. Cashmere is the product of secondary hair follicle with seasonal variation activity during the life cycle of goat, which grows rapidly in autumn and falls off in spring. Within a completely growth cycle, hair follicle underwent a process-oriented period of anagen to catagen and finally telogen. As one of the biggest goat-raising country all over the world, China has made great contribution to world animal fiber industry and acts indispensable role of world’s cashmere yield. For increasing the commercial value of Inner Mongolia cashmere goat, many researchers have paid attention to molecular biology recent years, tried to find out key functional genes which are associate with cashmere growth and establish molecular basis of cashmere features using cDNA library construction, SNP analysis, gene scanning and other molecular biology technical.
In recent years, additional research has further demonstrated that miRNA is associated with maintain embryo early multi-potency in mammalian, indicating that miRNA may also involve in the regulation of hair follicle activity during the goat life cycle. Hence, identification of goat novel miRNAs is a indispensable and meaningful study to disclose the molecular mechanism of production cashmere and mutton. To date, only 77 records of goat miRNAs on NCBI, which 69 records were identified by CHIP technology in goat skin from Zhang study [8]. The other 8 records were gain by searching known sheep miRNA homology sequence. The small amount of goat miRNAs due to the technical limitation such as CHIP technology has limitations to discover novel miRNAs and low accuracy and reproducibility and expensive cost and time-consuming for classic molecular biology approach, however, the whole genome of goat has not well mapped yet.
Here we establish comparative genomics approach combine bioinformatics prediction to identify novel miRNA and their potential targets in Cashmere goat base on evolutional conservation properties. 5 novel miRNAs (chi-miR-2284n, chi-miR-421*, chi-miR-421, chi-miR-1839 and chi-miR-374; Accession number: JQ002550–JQ002554) were identified by blasting known ruminant miRNA to goat UniGene data set. Further RT-PCR validation and miRNA expression pattern mapping monthly in the whole year showed all the 5 novel miRNAs expressed in goat skin and longissimus dorsi and chi-miR-1839, chi-miR-374b and chi-miR-2284 showed a significant higher expression than longissimus dorsi and reach crest in February, and trough in September and October. While chi-miR-421 and chi-miR-421*, reach their highest expression level in October in skin tissue, indicating these novel miRNAs are associate with regulating seasonal activity changes of goat hair follicle.
Results
5 novel miRNAs were identified belonged to 4 different families in goat skin and longissimus dorsi
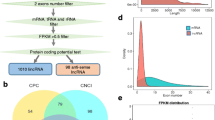
Base on the property of conservation among many species of miRNA and the feature of pre-miRNA secondary structure, we identified 5 novel miRNAs using homologue search with known miRNA sequence from other ruminants in goat UniGene data set. As shown in the Fig. 1, we name them as chi-miR-421/chi-miR-421*, chi-miR-374b, chi-miR-2284n and chi-miR-1839, respectively (Fig. 1a–d). The 5 pre-miRNAs were 64–80 nt in length, minimal folding free energy was less than −20 kcal/mol, σ_len was zero, (Table 1) all the parameters were in the set of threshold range and displayed highly conservative property among mammalian species. 5 novel miRNAs were belonging to 4 different miRNA families including a pair of miRNA/miRNA*. Notably, 4 miRNAs form a ~216 bp cluster, which was coincide with the previously report in human and mouse [9–11]. This cluster was located in the chromosome X, the 12th intron of non-coding gene Ftx in mouse, 8th intron in human and 9th intron in cow, respectively.
Validation of microRNAs expression with quantitative RT-PCR
To determine whether the identified miRNA express in goat, total RNA was prepared from longissimus dorsi, leg muscle and skin sample. The RT-PCR result demonstrates all the 5 novel miRNAs were expressed in goat skin and longissimus dorsi (Fig. 2a). Sanger sequencing further demonstrated that the 5 identified miRNAs were expressed in goat skin, longissimus dorsi and leg muscle.
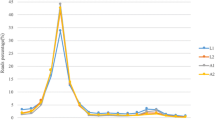
Validation of microRNAs expression by quantitative RT-PCR a Expression of chi-miR-421/chi-miR-421*, chi-miR-374b, chi-miR-1839 and chi-miR-2284n in skin, longissimus dorsi and skeletal muscle. b Overview of miRNAs expression pattern in the whole year in skin. c Expression pattern of chi-miR-374b in the whole year. d Expression pattern of chi-miR-1839 in the whole year. e Expression pattern of chi-miR-2284n in the whole year. f Expression pattern of chi-miR-421/chi-miR-421* in the whole year
Expression pattern during 12 months in the whole year
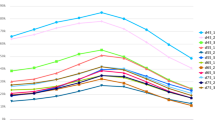
To explore whether these 5 novel miRNA contribute to the production of goat hair follicle activation, real time PCR was determined using classic standard curve method. By mapping the expression pattern of identified novel miRNA in the skin sample collected every month in the whole year, we eventually got the expression pattern monthly in the whole year. According to the real time PCR results, chi-miR-374b (Fig. 2c), chi-miR-1839 (Fig. 2d) and chi-miR-2284 (Fig. 2e) expressed with a similar expression pattern, it is significant higher expression than longissimus dorsi and reach crest in February, and trough in September and October. However, chi-miR-421 and chi-miR-421* expression were not parallel in the same tissue, chi-miR-421 expressed higher than chi-miR-421*, reach their highest expression level in October in skin tissue (Fig. 2f), indicating chi-miR-1839, chi-miR-374b and chi-miR-2284 play important role in pushing secondary hair follicle activity changes from catagen to telogen and chi-miR-421 and chi-miR-421* are involved in the molecular event of promoting cashmere growth spurt. Meanwhile, miRNA expression levels were determined in different muscle from male or female, adult or childhood goat. Compare with other miRNAs, chi-miR-374b showed significantly higher expression level in both male and female longissimus dorsi and hind leg muscle indicating that chi-miR-374b may contribute to muscle development (Fig. 3b).
MicroRNA target genes prediction and function analysis a the pattern of miRNA targets in skin b Expression pattern in muscle tissue. CMB, adult female longissimus dorsi. CGB, adult male longissimus dorsi. YMB, child female longissimus dorsi. YGB child male longissimus dorsi, CMH adult female hind leg, CGH adult male hind leg, YMH child female hind leg, YGH child male hind leg
MicroRNA target genes prediction and function analysis
To further determine the biological functions involved these miRNAs during the development of cashmere growth, target gene prediction was performed based on miRNA/mRNA interactions to provide some molecular insight into the processes. A PAGE electrophoresis combining sequencing approach was used to establish target genes map in goat skin and longissimus dorsi as Fig. 3a shown, in total 4 goat skin specific genes and 6 genes in longissimus dorsi were predicted as putative target genes for chi-miR-421* and chi-miR-1839 (Tables 2, 3). The predicted target genes of chi-miR-421* and chi-miR-1839, WDR12 and CSNK1A1 were annotation showed according to Notch/Wnt single transduction pathway, which are not only important for stem cell proliferation and differentiation but also participate in early development and terminal differentiation of hair follicle. The other 2 goat specific target genes in skin of chi-miR-421* Unigene29624_NMCG and Unigene70278_NMCGR are not well annotated yet. Target gene PIP5K1A of chi-miR-421* in longissimus dorsi was involved in metabolic pathway and ELMOD2 of chi-miR-374b, EIF3F of chi-miR-421 were related to the functions of TLR3 pathway and RNA transport, respectively.
Material and method
Tissue sample
Inner Mongolia white cashmere goats were provided by livestock farms in Otog Banner, Ordos City, Inner Mongolia Autonomous Region, China. The feeding and management conditions are quite the same for all the animals. Sample collection of longissimus dorsi, leg and skin tissue was done under a completely growth cycle, which from July on 2008 to 2009, 12 months in total. Longissimus dorsi and leg muscle were collected from 12 cashmere white goat (3 adult males, 3 adult females, 3 lamb males and 3 lamb females), Skin tissue got from 3 female cashmere white goat with 24.5 kg weight on average, 1 year old every month. 1 cm2 skin samples were collected from the side body of goats, and then frozen in liquid nitrogen and stored at −80 °C for further analysis. Longissimus dorsi and leg samples were collected while the animals were slaughtered. All animal experiments were performed in accordance with the “Guidelines for Experimental Animals” of the Ministry of Science and Technology (Beijing, China). All surgery was performed according to recommendations proposed by the European Commission (1997), and all efforts were made to minimize suffering.
Published data acquisition
UniGene data set was kindly provided by Inner Mongolia ATCG institute of bioinformatics, totally 82,582 Expressed Sequence Tag. Published miRNA and pre-miRNA sequence (totally 15,172 miRNAs/pro-miRNAs) were downloaded from miRBase (http://www.mirbase.org/ftp.shtml;2011.1.10).
RNA extraction and RT-PCR
Total RNA was isolated and then reverse transcribed to cDNA using the TRIzol reagent and the reverse transcription system, respectively, according to manufacturer’s instructions. Triplication gene expression assays were performed by using SYBR Green qPCR Master Mix on a MyiQThermocycler (Bio-Rad). All transcript levels were normalized to 5S rRNA. (See Table.. for primer sequences).
Novel microRNA prediction
In this study, we researched novel miRNA sequence base on conservation property of miRNA, pre-miRNA and secondary structure using homology fragment blasting in goat UniGene database. Follow the procedure and criteria as below: (1) gate out redundancy miRNA sequence (2) blast non redundancy miRNA to goat UniGene, E = 10 (3) blast UniGene sequence which false matching base number ≤2 to downloaded pre-miRNA sequence. E = 10 (4) wipe off redundancy of pre-miRNA sequence which consensus rate more than 90 % (5) wipe protein coding sequence (6) get a rough result (7) use Mfold software to simulate pre- miRNA secondary structure as following criteria: (1) A + U content between 30 to 70 % (2) mature miRNA produce from one arm with same position of homology sequence in different species (δ-len [7], δ-len = 10 as threshold) [12]; (3) minimum free energy (MFE)of pre-miRNA must less than −20 kcal/mol; (4) at least 16 bp mature miRNA located in stem region, no large and nonasymmetric structure [13].
miRNA clone and sequencing
Monthly expressed miRNAs in goat skin were reverse transcripted using oligo (dT) as primer, and then ligate into pGM-T vector for Sanger sequencing.
Discussion
In present study, we identified 5 novel microRNAs expressed in goat skin and longissimus dorsi, which a cluster composed of chi-miR-421, chi-miR-421* and chi-miR-374b located on chromosome X. We determined the A/U content threshold of pre-miRNA to 30–80 % refer to Sheng [12] and Xie’s [13] research and other published miRNA information. Sequence analysis demonstrated that chi-miR-2284n was a member of cow specific miR-2284 family [14] and differed from bta-miR-2284 l which was derivate from different precursors. Collectively, we conclude that chi-miR-2284n functions as ruminant specific miRNA in cow, goat and etc.
According to the result of real time PCR, we found that chi-miR-1839, chi-miR-374b and chi-miR-2284n showed a significant high expression in skin with the highest expression level in February. Goat secondary hair follicle goes into telogen from anagen and catagen in February, while lost the ability of producing cashmere. We conclude that some molecular events trigger the expression of chi-miR-1839, chi-miR-374b and chi-miR-2284n and subsequently repress their targets to promote goat hair follicle growth from catagen to telogen. In contrast, chi-miR-421 and chi-miR-421* expressed both in skin with a similar expression pattern and reach a highest expression level in October when cashmere grows rapidly. Thus chi-miR-421 and chi-miR-421* may acts to promote cashmere production. The changes of expression level as hair follicle activity demonstrated the association of miRNA expression with cashmere production. As the secondary product of cashmere goat breeding industry, meat production was paid attention by researchers as well. In present study, we also determined the expression of these 5 novel identified miRNAs in longissimus dorsi and skeletal muscle. All the 5 miRNAs were expressed in longissimus dorsi and skeletal muscle, with the similar expression pattern in the whole year in skin. It indicated these miRNAs not only regulated cashmere production but also meat production. Further exploration of potential target genes of these miRNA showed that Notch/Wnt single transduction pathway was involved in this molecular process as reported by other researchers in stem cell regulation. Our results demonstrated that the important pathways in development like Notch/Wnt single transduction pathway were also finely regulated by miRNAs in cashmere goat.
To get high quality functional miRNA in the biology event of goat cashmere production, we use a strict parameter setting to screen novel miRNA, With relatively lower efficiency compared with the study of Zhang [8] and Cheng [15], our study identified only 5 novel miRNAs in totally 82,580 expression sequence taqs, That is mainly because of our strict parameter setting and the technical bias and limitation using oligo dT when reverse transcription so that the miRNAs spliced from intron could not be detected.
References
Lee RC, Feinbaum RL, Ambros V, The C (1993) Elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75(5):843–854
Ambros V (2004) The functions of animal microRNAs. Nature 431(7006):350–355
Chen K, Rajewsky N (2007) The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet 8(2):93–103
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136(2):215–233
Xihui S, Lixin D (2007) Progress on the research of microRNAs and its function in humans and animals. Hereditas (Beijing) 29(6):651–658
Kusenda B et al (2006) MicroRNA biogenesis, functionality and cancer relevance. Biomed Papers 150(2):205–215
Saini HK, Griffiths-Jones S, Enright AJ (2007) Genomic analysis of human microRNA transcripts. Proc Natl Acad Sci 104(45):17719–17724
Wenguang Z et al (2007) A subset of skin-expressed microRNAs with possible roles in goat and sheep hair growth based on expression profiling of mammalian microRNAs. Omics 11(4):385–396
Chureau C et al (2011) Ftx is a non-coding RNA which affects Xist expression and chromatin structure within the X-inactivation center region. Hum Mol Genet 20(4):705–718
Altuvia Y et al (2005) Clustering and conservation patterns of human microRNAs. Nucleic Acids Res 33(8):2697–2706
He P-A et al (2008) Identification and characteristics of microRNAs from Bombyx mori. BMC Genom 9(1):248
Sheng X et al (2011) Characterization of microRNAs from sheep (Ovis aries) using computational and experimental analyses. Mol Biol Rep 38(5):3161–3171
Xie FL et al (2007) Computational identification of novel microRNAs and targets in Brassica napus. FEBS Lett 581(7):1464
Borchert GM et al (2011) Comprehensive analysis of microRNA genomic loci identifies pervasive repetitive-element origins. Mob Genet Elem 1(1):8–17
Haixuan C et al (2008) Computational and experimental identification of novel microRNAs in goat. Hereditas (Beijing) 30(10):1326–1332
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Su, R., Fu, S., Zhang, Y. et al. Comparative genomic approach reveals novel conserved microRNAs in Inner Mongolia cashmere goat skin and longissimus dorsi. Mol Biol Rep 42, 989–995 (2015). https://doi.org/10.1007/s11033-014-3835-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3835-9