Abstract
Preadipocyte factor 1 (Pref-1), also known as a delta-like 1 protein, is a transmembrane and secreted protein containing the epidermal growth factor-like repeat. Pref-1 inhibits adipocyte differentiation by activating the ERK1/2 pathway. MicroRNAs, a new class of small noncoding RNAs of 20–24 nucleotides, act as negative regulators of gene expression and result in mRNA degradation or translational repression. MicroRNA-143 (miR-143) is known to induce adipocyte differentiation; however, miR-143 targets in the regulation of adipocyte differentiation remain unknown. In this study, we investigated whether pref-1 is a miR-143 target to regulate adipogenesis. After the induction of adipocyte differentiation the level of miR-143 was increased, whereas the expression of pref-1 mRNA was decreased. The pref-1 protein level was also down-regulated in preadipocytes ectopically expressing miR-143, and recovered by miR-143 inhibitor. The binding region for miR-143 was predicted to be located between positions 247 and 252 in the 3′-UTR of pref-1. The luciferase activity of the vector containing the wild-type 3′-UTR of pref-1 was decreased by 65 % in cells transfected with miR-143 mimic compared to that of the corresponding control. In contrast, the activity of the pref-1 mutant cells was not affected by the treatment with miR-143 mimic. The ectopic expression of miR-143 mimic suppressed the phosphorylation of ERK1/2 induced by pref-1 in 3T3-L1 cells. However, the suppressed phosphorylation was restored by miR-143 inhibitor. Taken together, these data suggest that miR-143 promotes adipogenesis by directly modulating the pref-1 expression in adipocytes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Adipocyte differentiation is the result of transcriptional remodeling leading to the activation of a large number of adipose-related genes. Preadipocyte factor 1 (Pref-1), a member of epidermal growth factor-like repeat family, is a transmembrane and secreted protein. Pref-1 plays an important role in the regulation of adipocyte differentiation [1, 2]. Since pref-1 is highly expressed in preadipocyte but not in mature adipocyte, it is used as a preadipocyte marker in adipose tissue [3]. Overexpression of pref-1 in mice is shown to reduce adipose tissue mass [4]. Pref-1 activates the MEK/ERK pathway to inhibit adipocyte differentiation [5]; therefore, repression of pref-1 is required for the induction of adipocyte differentiation. However, the molecular regulation of pref-1 expression during adipogenesis is not yet fully understood.
MicroRNAs (miRNAs) are endogenous noncoding RNAs of 20–24 nucleotides in length and play an important role in the negative regulation of gene expression by base-pairing to the complementary site on the target mRNAs, which in turn causes an inhibition of translation or the degradation of target mRNAs [6]. The role of miRNAs in adipogenesis is first found by a study of miR-143, which induces adipogenesis in adipocytes [7]. Further studies showed that several miRNAs, including the miR-17/92 cluster, miR-103, and miR-125a, were also upregulated in mature adipocytes [8, 9]. Recently, miRNA profiling studies have identified novel miRNAs that are involved in adipogenesis and associated with obesity [10–12]; however, the challenge remains to determine the mechanism how these miRNAs regulate their targets in adipose tissue. Therefore, in this study, we demonstrated that miR-143 regulates the pref-1 expression during adipogenesis.
Materials and methods
Materials
The pGL3-control firefly luciferase vector, the pRL-TK Renilla luciferase vector, and the dual luciferase reporter assay system were purchased from Promega (Madison, WI, USA). A mouse 3T3-L1 cell line was obtained from American Type Culture Collection (Rockville, MD, USA). Dulbecco’s modified Eagle’s medium (DMEM), calf serum and fetal bovine serum (FBS) were acquired from Gibco/BRL (Grand Island, NY, USA). Lipofectamine 2000 transfection reagents and oligofectamine reagents were obtained from Invitrogen (Carlsbad, CA, USA). 3-Isobutyl-l-methylxanthine (MIX), dexamethasone (DEX), and insulin were purchased from Sigma (St. Louis, MO, USA). The QuikChange site-directed mutagenesis kit was obtained from Stratagene (La Jolla, CA, USA).
Plasmid constructs
The pGL3-control vector was used to construct pGL3-pref-1 wt, which contained the 3′-UTR of mouse pref-1 (GenBank accession No. NM003836). Briefly, DNA fragments generated by polymerase chain reaction (PCR) were digested with the restriction enzyme XbaI to generate appropriate protruding ends. Site-directed mutagenesis of the pref-1 3′-UTR was performed using the QuikChange site-directed mutagenesis kit following the supplier’s instructions. The original sequence TCATCTC in the miR-143-binding site of the pref-1 3′-UTR was mutated to TCAGACC. Successful mutagenesis was confirmed by the sequence analysis.
Cell culture
3T3-L1 preadipocytes cultured in DMEM supplemented with 10 % calf serum were allowed to reach confluence. Two days after reaching confluence (designed as day 0), the differentiation of preadipocytes was initiated by adding 10 µg/ml insulin, 1 µM DEX, and 0.5 mM MIX in DMEM supplemented with 10 % FBS. After 48 h (day 2), culture media were replaced with DMEM supplemented with 10 % FBS and 1 µg/ml insulin, and cells were then fed every other day. By day 8, cells were fully differentiated, and cytoplasmic triglyceride droplets were visible.
Real-time reverse transcription-PCR
Total RNA was isolated from 3T3-L1 cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The synthesis of cDNA was performed using MMLV reverse transcriptase (Takara, Shinga, Japan) and oligo dT primers (Invitrogen, Carlsbad, CA, USA). Real-time quantitation was performed using the iCycleriQ system (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s protocol. The fluorescence threshold value was calculated using the iCycleiQ system software. Reverse transcription reaction mixtures were incubated with iQ SYBR Green Supermix (Bio-Rad, Hercules, CA, USA). Data processed by the comparative cycle threshold method were expressed as fold increases relative to the basal transcription level. The amount of target mRNA was normalized to GAPDH mRNA. Oligonucleotides used in this study were as follows: pref-1, 5′-TGG CTT CTC AGG CAA CTT CT-3′ and 5′-CTT GCA CAG ACA CTC GAA GC-3′; GAPDH, 5′-GAC TTC AAC AGC AAC TCC CAC-3′ and 5′-TCC ACC ACC CTG TTG CTG TA-3′.
For quantitative RT-PCR of microRNAs, total RNA was extracted using TRIzol reagent as described before and was processed reverse transcription. The cDNA products were used for PCR. Taqman primers and probes for miR-143 were purchased from Applied Biosystems (Foster City, CA, USA). The amplification and detection of miR-143 were performed using the iCycleriQ system with 40 cycles of denaturation at 95 °C for 15 s and annealing/extension at 60 °C for 60 s. U6 snRNA was used as the internal control. Real-time PCR was performed in triplicate. Data were analyzed by the relative quantification (ΔΔC t) method.
Transient transfection and luciferase reporter assays
For transient miRNA transfection, 3T3-L1 cells were transfected with miR-143 mimic, miR-143 inhibitor (GenePharma, Shanghai, China), or scrambled oligonucleotides as a control using oligofectamine reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s instructions. For luciferase report assay COS7 cells were co-transfected with reporter plasmids in combination with either pcDNA3.1 or pcDNA3.1-miR-143 plasmids. Luciferase activities were measured at 48 h post-transfection using the dual-luciferase assay system (Promega, Madison, WI, USA) with a GloMax20/20 luminometer (Turner BioSystem, Sunnyvale, CA, USA).
Western blot analysis
Cells were lysed in ice-cold lysis buffer [50 mM Tris–HCl, 150 mM NaCl, 1 % NP-40, 0.1 % SDS, protease inhibitor cocktail (Roche, Indianapolis, IN, USA), 50 mM NaF, and 0.2 M Na3VO4]. Protein extracts were separated by SDS-PAGE and blotted onto nitrocellulose transfer membranes. Blocking was performed at room temperature for 1 h in TBS-T with 5 % BSA, followed by incubation with anti-ERK1/2 (Cell Signaling Technology, Beverly, MA, USA), anti-phospho-ERK1/2 (Cell Signaling Technology, Beverly, MA, USA), anti-pref-1/dlk1 (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA), or anti-actin (Sigma, St. Louis, MO, USA) antibodies in TBS-T. After washes with PBS, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies. Protein signals were then visualized using an enhanced chemiluminescence kit (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA). Intensity of each protein band was quantified by densitometry.
Bioinformatics
The analysis of miR-143 predicted targets was determined using the algorithms of TargetScan 6.2 (http://www.targetscan.org) and miRanda (http://www.microrna.org). According to these algorithms, microRNA with the highest mirSVR score was selected for further study.
Statistics
Data are presented as mean ± SE of three independent experiments. The statistical significance of between two groups was assessed using the Student t test. All statistical analysis was carried out using SPSS version 11.0 software (SPSS, Chicago, IL, USA).
Results
The expression of pref-1 and miR-143 in 3T3-L1 cells during adipocyte differentiation
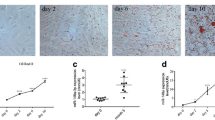
To determine target microRNAs of pref-1, in silico algorithms (miRanda and TargetScan program) were used; as a result, miR-143 was predicted to be a regulator of pref-1 because miR-143 had the highest mirSVR score among candidate genes. We next investigated the expression of both miR-143 and pref-1 in 3T3-L1cells before and after adipocyte differentiation. As shown in Fig. 1a, the level of pref-1 mRNA was strongly expressed in preadipocytes compared to mature adipocytes. On day 8 after adipocyte differentiation, the expression of pref-1 mRNA was almost undetectable, whereas the level of miR-143 mRNA was increased exponentially. The protein level of pref-1 in mature adipocytes was also decreased gradually by day 8 post-differentiation (Fig. 1b). These results suggest that the expression of both genes may be correlated with the differentiation stage of adipocytes.
Expression of pref-1 and miR-143 in 3T3-L1 cells during adipogenesis. Preadipocyte cells were stimulated to undergo adipocyte differentiation at 2 days post-confluence. a After the induction of differentiation total RNA prepared from 3T3-L1 cells at indicated time points was subjected to quantitative real-time RT-PCR. The amount of pref-1 mRNA and miR-143 were normalized to GAPDH mRNA and U6 snRNA, respectively. Data shown represent fold changes of mRNA or miRNA levels between day 0 and day 8 after the induction of differentiation. Results are expressed as mean ± SE; n = 3. * p < 0.05, ** p < 0.01. b The pref-1 protein levels were measured by western blotting at indicated time points after adipocyte differentiation. Actin was used as a loading control. The graph represents the quantification of pref-1 protein resulting from western blot data. Experiments were performed in triplicate and representative results are shown
Suppression of pref-1 gene expression in cells transfected with miR-143
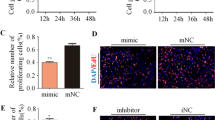
To investigate whether miR-143 regulates the pref-1 expression, miR-143 mimic was transfected into 3T3-L1 cells, and the pref-1 mRNA expression was then measured. As a result, the pref-1 mRNA level in miR-143-overexpressing cells was reduced by 50 % compared to the control (Fig. 2a). Similarly, the pref-1 protein level was also decreased by 60 % in cells exposed to miR-143 mimic, whereas miR-143 inhibitor recovered dose-dependently the down-regulated pref-1 expression in 3T3-L1 cells (Fig. 2b). In mouse primary pre-adipocytes, the level of pref-1 expression was down-regulated by miR-143 mimic (Fig. Suppl a and b). These results suggest that miR-143 regulates the pref-1 gene expression in adipocytes.
Effects of miR-143 overexpression on the pref-1 expression in 3T3-L1 cells. a Cells were transfected with 50 nM miR-143 mimic, and then incubated for 48 h. The pref-1 expression was measured by quantitative real-time RT-PCR. The amount of pref-1 mRNA was normalized to GAPDH mRNA. Results are expressed as mean ± SE of three independent experiments. * p < 0.05. b The pref-1 protein levels in cells treated with miR-143 or miR-143 inhibitor were measured by western blotting at 48 h post-transfection. The graph indicates the band intensities of pref-1 normalized to actin. Representative blots from three independent experiments are shown. NC negative control Inh inhibitor
Identification of miR-143 target sites in the 3′- UTR of pref-1
Based on the expression study of both genes described above, the TargetScan program was used to predict miR-143-binding sites in the 3′-UTR region of pref-1. As shown in Fig. 3a, the region between positions 247 and 252 of the pref-1 3′-UTR was expected to be a seed match of miR-143. Subsequently, this putative miR-143-interacting region in the pref-1 3′-UTR was constructed into reporter plasmids and the potential binding site was also mutated by site-directed mutagenesis (Fig. 3b). In the presence of miR-143 mimic, the luciferase activity was measured in COS7 cells transfected with these reporter plasmids. The activity of the reporter plasmid containing the wild-type 3′-UTR of pref-1 was decreased by 65 % compared to that of the control (Fig. 3c). However, the activity of the mutant construct was almost similar to the control in miR-143-overexpressing cells. These results indicate that miR-143 binds to the pref-1 3′-UTR to regulate the post-transcriptional expression of pref-1.
The interaction of miR-143 with the pref-1 3′-UTR. a The putative miR-143-binding site in the 3′-UTR of mouse pref-1 is matched with the miR-143 seed region. b Schematic representation of the reporter vector carrying the pref-1 3′-UTR. The 3′-UTR region of pref-1 was cloned into the downstream of the firefly luciferase gene (Luc+) in the reporter vector pGL3 (pGL3-pref-1 wt), and a mutant construct was obtained by changing 3 base pairs of the miR-143 seed sequence (pGL3-pref-1 mu). c COS7 cells were co-transfected with a reporter vector (pGL3-control, pGL3-pref-1 wt, or pGL3-pref-1 mu) and the pcDNA-miR-143 expression vector. Luciferase activities were measured using the dual-luciferase reporter assay at 48 h post-transfection and then normalized to the Renilla luciferase. Results are presented as mean ± SE of three independent experiments. ** P < 0.01
Reduced functional activity of pref-1 in response to miR-143
To determine whether the reduced expression of pref-1 is dependent on miR-143 during adipogenesis, the pref-1 mRNA level was measured in mature adipocytes exposed to the miR-143 inhibitor (anti-miR-143). After transfection, the cells were incubated with growth medium or differentiation medium and the pref-1 level was examined 2 days later. As we expected, the cells grown in growth medium expressed high levels of pref-1 mRNA, whereas those grown in differentiation medium showed decreased pref-1 expression (Fig. 4a). The pref-1 expression affected by the miR-143 inhibitor in the cells grown in growth medium was negligible, consistent with the fact that the miR-143 expression in preadipocytes is weak, and therefore the effect of miR-143 inhibitor is thought to be little. In contrast, during the differentiation process, the miR-143 inhibitor was found to increase the pref-1 expression in the cells grown in differentiation medium (Fig. 4a).
Suppression of ERK1/2 phosphorylation by miR-143 in adipocytes. a 3T3-L1 adipocyte cells were transfected with either 50 nM miR-143 inhibitor (anti-miR-143) or scrambled control. At 48 h post-transfection, cells were incubated in growth medium (GM) or differentiation medium (DM) for 2 days. Total RNA was isolated from cell lysates and subjected to RT-PCR. b Cells were transfected with 50 nM miR-143 mimic, miR-143 inhibitor or scrambled control (NC), followed by the induction of differentiation. Cell lysates were collected and subjected to western blotting. The graph represents the band intensities of ERK1/2 phosphorylation normalized to total ERK1/2 protein level. Representative results are shown from three independent experiments. Inh inhibitor
The inhibition of adipocyte differentiation by pref-1 is known to induce the phosphorylation of ERK1/2 [5]. To investigate the function of miR-143 in adipocytes, the level of ERK1/2 phosphorylation was measured after the cells were transfected with miR-143 mimic. As shown in Fig. 4b and Fig Suppl c, ERK1/2 phosphorylation in miR-143-overexpressing cells was reduced by 50 % compared to the control, even though the total ERK1/2 protein level was not changed. However, in the presence of miR-143 inhibitor, the phosphorylation of ERK1/2 was dose-dependently restored in the miR-143 overexpressing cells (Fig. 4b). Taken together, these results suggest that miR-143 plays a role as a negative regulator of pref-1 expression during adipocyte differentiation.
Discussion
Adipocyte differentiation is occurred by systemically coordinated gene expressions [13]. Although PPARγ and C/EBPα have been identified as key regulators in adipogenesis, the detailed molecular mechanism including microRNAs remains to be fully elucidated. MicroRNAs exert their actions primarily at the post-transcriptional level via translational repressor and/or mRNA degradation [6, 14]. Some microRNAs such as miR-17/92 cluster, miR-103, and miR-143 are known to enhance adipocyte differentiation [9, 15]. It is well known that miR-143 is up-regulated after the induction of adipocyte differentiation. MiR-143 inhibitor induces the suppression of adipocyte differentiation, reduces the storage of fat droplets, and decreases the expression of key adipogenic genes such as CEBPα and FABP4 [16]. In this study, we analyzed the mechanism by which miR-143 involves the transition of 3T3-L1 preadipocytes to adipocytes. One of the miR-143 targets is reported as ERK5 in human adipocytes [7]. ERK5 is known to promote cell growth and proliferation in response to tyrosine kinase signaling [7, 17]. Similar results were obtained in our study where miR-143 suppressed ERK1/2 phosphorylation in 3T3-L1 cells via downregulated pref-1 level. The ERK activation phosphorylates PPARγ and C/EBP, thereby inducing a decrease in PPARγ transcriptional activity [18]. Pref-1 induces the phosphorylation of ERK1/2 and in turn inhibits adipocyte differentiation [5]. The MAPK signaling pathways activate a variety of genes involved in adipocyte growth and differentiation [19, 20]. The activation of ERKs in the process of adipocyte differentiation is tightly and temporally controlled, and depends on many parameters, since under certain conditions ERK activity may be required for adipogenesis, while in other conditions ERK activity may impair adipocyte differentiation [21, 22]. In the terminal differentiation stage of adipocyte, the activation of ERKs inhibits cell differentiation via the phosphorylation of PPARγ [23]. The upstream kinase that directly phosphorylates ERK5 is identified as MAP2K5 [23, 24], which is a direct target of miR-143. During the terminal differentiation stages, the overexpression of miR-143 blocks the MAP2K5-ERK5 and ERK1/2 signaling pathways, so the ERK-mediated phosphorylation of PPARγ is reduced. Further studies are required to determine the molecular mechanism of the miR-143-mediated MAPK signaling pathway in adipocyte differentiation.
In the present study, we showed the expression pattern of pref-1 level in 3T3-L1 cells. Pref-1 was strongly expressed in preadipocytes but drastically reduced at day 8 after the induction of differentiation. To date, the pref-1 expression is reported to be transcriptionally regulated by the Rb-associated E2F1 complex via the phosphorylation of Rb [25, 26]. Interestingly, during adipogenesis the reduced expression of E2F1 is accompanied by the diminished level of pref-1. However, at late stage of adipocyte differentiation the expression patterns of E2F1 and pref-1 are inconsistent; this is, the E2F1 expression is increased, whereas the level of pref-1 expression is decreased. These discordant observations indicate that other mechanisms may exist to regulate the pref-1 expression. One of regulatory mechanisms for the pref-1 stability is microRNA process, which regulates post-transcriptional gene expression. Some microRNAs play a role in modulating adipocyte differentiation. In previous study, miR-15 is reported to regulate the expression of pref-1, even though the direct interaction between pref-1 and miR-15 is not showed [27]. In this study, we reported that miR-143 was expressed more strongly at day 8 after differentiation than preadipocytes and affected the expression of pref-1 by binding its 3′-UTR region. Consistent with our study, it is reported that miR-143 is predominantly expressed at late stage of differentiation [15]. These results indicate that the level of pref-1 expression at the late terminal differentiation stage is regulated by miR-143. Furthermore, by using in silico TargetScan miRNA prediction program (version 6.2), 23 putative target miRNAs against mouse pref-1/dlk-1 were found a conserved binding site in the 3′-UTR of pref-1, leading to the putative repression of pref-1 level by several different microRNAs. Therefore, further analysis is necessary to elucidate the regulatory mechanism of pref-1 expression in adipocyte differentiation.
In conclusion, our data suggest that miR-143 promotes the ERK-mediated adipocyte differentiation by modulating the adipogenic inhibitor pref-1 in 3T3-L1 cells. Based on our study, the molecular mechanism of miR-143 may contribute to the prevention of obesity and its related diseases.
References
Smas CM, Sul HS (1993) Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell 73:725–734
Ansell PJ, Zhou Y, Schjeide BM, Kerner A, Zhao J, Zhang X, Klibanski A (2007) Regulation of growth hormone expression by Delta-like protein 1 (Dlk1). Mol Cell Endocrinol 271:55–63
Lee K, Villena JA, Moon YS, Kim KH, Lee S, Kang C, Sul HS (2003) Inhibition of adipogenesis and development of glucose intolerance by soluble preadipocyte factor-1 (Pref-1). J Clin Invest 111:453–461
Villena JA, Choi CS, Wang Y, Kim S, Hwang YJ, Kim YB, Cline G et al (2008) Resistance to high-fat diet-induced obesity but exacerbated insulin resistance in mice overexpressing preadipocyte factor-1 (Pref-1): a new model of partial lipodystrophy. Diabetes 57:3258–3266
Kim KA, Kim JH, Wang Y, Sul HS (2007) Pref-1 (preadipocyte factor 1) activates the MEK/extracellular signal-regulated kinase pathway to inhibit adipocyte differentiation. Mol Cell Biol 27:2294–2308
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Esau C, Kang X, Persalta E, Hanson E, Maarcusson EG, Ravichandran LV et al (2004) MicroRNA-143 regulates adipocyte differentiation. J Biol Chem 279:52361–52365
Wilfred BR, Wang WX, Nelson PT (2007) Energizing miRNA research: a review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol Genet Metab 91:209–217
Xie H, Lim B, Lodish HF (2009) MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes 58:1050–1057
Bengestrate L, Virtue S, Campbell M, Vidal-Puig A, Hadaschik D, Hahn P, Bielke W (2011) Genome-wide profiling of microRNAs in adipose mesenchymal stem cell differentiation and mouse models of obesity. PLoS One 6:e21305
Keller P, Gburcik V, Petrovic N, Gallaqher IJ, Nederqaard J, Cannon B, Timmons JA (2011) Gene-chip studies of adopogenesis-regulated microRNAs in mouse primary adipocytes and human obesity. BMC Endocr Disord 22:7
Chartoumpekis DV, Zaravinos A, Ziros PG, Iskrenova RP, Psyrogiannis AI et al (2012) Differential expression of microRNAs in adipose tissue after long-term high—fat diet-induced obesity in mice. PLoS One 7:e34872
Farmer SR (2006) Transcriptional control of adipocyte formation. Cell Metab 4:263–273
Kim JH, Park SG, Song SY, Kim JK, Sung JH (2013) Reactive oxygen species-responsive miR-210 regulates proliferation and migration of adipose-derived stem cells via PTPN2. Cell Death Dis 4:e588
Kajimoto K, Naraba H, Iwai N (2006) MicroRNA and 3T3-L1 pre-adipocyte differentiation. RNA 12:1626–1632
Li H, Zhang Z, Zhou X, Wang Z, Wang G, Han Z (2011) Effects of microRNA-143 in the differentiation and proliferation of bovine intramuscular preadipocytes. Mol Biol Rep 7:4273–4280
Kato Y, Tapping RI, Huang S, Watson MH, Ulevitch RJ, Lee JD (1998) Bmk1/Erk5 is required for cell proliferation induced by epidermal growth factor. Nature 395:713–716
Prusty D, Park BH, Davis KE, Farmer SR (2002) Activation of MEK/ERK signaling promotes adipogenesis by enhancing peroxisome proliferator-activated receptor gamma (PPAR gamma) and C/EBP alpha gene expression during the differentiation of 3T3-L1 preadipocytes. J Biol Chem 277:46226–46232
Seger R, Krebs EG (1995) The MAPK signaling cascade. FASEB J 9:726–735
Gwon SY, Ahn JY, Jung CH, Moon BK, Ha TY (2013) Shikonin suppresses ERK1/2 phosphorylation during the early stages of adipocyte differentiation in 3T3-L1 cells. BMC Complement Altern Med 13:207
Chen L, Hou J, Ye L, Chen Y, Cui J et al (2014) MicroRNA-143 regulates adipogenesis by modulating the MAP2K5-ERK5 signaling. Sci Rep 4:3819
Ling HY, Wen GB, Feng SD, Tuo QH, Ou HS et al (2011) MiRNA-375 promotes 3T3-L1 adipocyte differentiation via modulation of ERK signaling. Clin Exp Pharmacol Physiol 38:239–246
Hu E, Kim JB, Sarraf P, Spiegelman BM (1996) Inhibition of adipogenesis through MAP kinase-mediated phosphorylation of PPAR gamma. Science 274:100–103
Zhou G, Bao ZQ, Dixon JE (1995) Components of a new human protein kinase signal transduction pathway. J Biol Chem 270:12665–12669
Fajas L, Landsberg RL, Huss-Garcia Y, Sardet C, Lees JA, Auwerx J (2002) E2Fs regulate adipocyte differentiation. Dev Cell 3:39–49
Shen YN, Kim YM, Yun CH, Moon YS, Kim SH (2009) Transcriptional activation of pref-1 by E2F1 in 3T3 L1 cells. BMB Rep 42:691–696
Andersen DC, Jensen CH, Schneider M, Nossent AY, Eskildsen T, Hansen JL, Teisner B, Sheikh SP (2010) MicroRNA-15a fine-tunes the level of Delta-like 1 homolog (DLK1) in proliferating 3T3-L1 preadipocytes. Exp Cell Res 316:1681–1691
Acknowledgments
This research was supported by grants from Basic Science Research Program through the National Research Foundation of Korea (NRF) (No. 2012R1A1B3001134), and by the Next-Generation BioGreen 21 Program (No. PJ008116), Rural Development Administration (RDA), Korea.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kim, YJ., Min, T.S., Seo, KS. et al. Expression of pref-1/dlk-1 is regulated by microRNA-143 in 3T3-L1 cells. Mol Biol Rep 42, 617–624 (2015). https://doi.org/10.1007/s11033-014-3807-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3807-0