Abstract
Myotonic dystrophy type 1 (DM1) is a multisystem genetic disorder caused by a triplet nucleotide repeat expansion in the 3′ untranslated region of the Dystrophia Myotonica-Protein Kinase (DMPK) gene. DMPK gene transcripts containing CUG expanded repeats accumulate in nuclear foci and ultimately cause altered splicing/gene expression of numerous secondary genes. The study of primary cell cultures derived from patients with DM1 has allowed the identification and further characterization of molecular mechanisms underlying the pathology in the natural context of the disease. In this study we show for the first time impaired nuclear structure in fibroblasts of DM1 patients. DM1-derived fibroblasts exhibited altered localization of the nuclear envelope (NE) proteins emerin and lamins A/C and B1 with concomitant increased size and altered shape of nuclei. Abnormal NE organization is more common in DM1 fibroblasts containing abundant nuclear foci, implying expression of the expanded RNA as determinant of nuclear defects. That transient expression of the DMPK 3′ UTR containing 960 CTG but not with the 3′ UTR lacking CTG repeats is sufficient to generate NE disruption in normal fibroblasts confirms the direct impact of mutant RNA on NE architecture. We also evidence nucleoli distortion in DM1 fibroblasts by immunostaining of the nucleolar protein fibrillarin, implying a broader effect of the mutant RNA on nuclear structure. In summary, these findings reveal that NE disruption, a hallmark of laminopathy disorders, is a novel characteristic of DM1.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Myotonic dystrophy type 1 (DM1) is an autosomal dominant inherited disease that represents the most common form of muscular dystrophy in adults, with a prevalence of 1 in 8,000 individuals worldwide [1]. DM1 is characterized mainly by myotonia (sustained muscle contraction), progressive muscle weakness and wasting as well as variable multisystemic features, including insulin resistance, cardiac conduction defects, gonadal atrophy, early posterior iridescent cataracts, and central nervous alterations [1, 2]. DM1 is caused by an expanded CTG repeat in the 3′-untranslated region (3′ UTR) of the DMPK gene in chromosome 19q [3–5]. Because of the location of CTG repeats in the 3′ UTR, it has been difficult to explain how a mutation in a non-coding region could cause the multisystemic features of DM1.
DM1 appears to be caused by different molecular mechanisms, including DMPK haploinsufficiency and a toxic gain-of-function by the expanded CUG repeat in mutant DMPK mRNA (reviewed in [6, 7]). Due to decreased levels of mRNA and protein of DMPK gene in adult DM1 tissue [8], the functional implications of a reduction in DMPK gene expression were genetically tested by generation of DMPK gene knockout mice [9]. However, Dmpk −/− mice exhibited only a mild late-onset, progressive skeletal myopathy with some cardiac- conduction abnormalities [10], implying that simply DMPK deficiency does not explain the multi systemic features of DM1. The latter hypothesis proposes that mutant DMPK mRNA containing expanded CUG repeats accumulates in the nucleus and folds into RNA hairpins, trapping alternative splicing factors, including muscleblind-like splicing regulator 1 (MBNL1) and hnRNP H, and activating the alternative splicing factor CUG triplet repeat RNA-binding protein 1 (CUG-BP1), also denominated CUGBP1/Elav-like family member 1 (CELF1), through hyperphosphorylation and stabilization in the cell nucleus [11–15], which in turns causes interference in developmentally regulated alternative splicing of defined pre-mRNAs [16–20]. In addition, mutant DMPK mRNA also binds and sequesters transcription factors, including Sp1 (specific protein 1) and STAT1 and 3 (members of the signal transducer and activator of transcription family), leading to reduce transcription of selected genes [19]. Furthermore, it has recently shown that CTG-repeat expansion induces repressive changes in chromatin dynamics [21].
In spite of the fact that expression of the CUG expanded repeats-containing mRNA interferes with different nuclear processes, and of the compelling evidence showing functional interrelationship between nuclear structure and function [22], the impact of this pathological transcript on nuclear architecture is largely unknown. In this study we show for the first time that expression of DM1-associated CUG repeats impairs the organization of different nuclear compartments, including the nuclear envelope (NE) and nucleoli, which results in nuclear morphology alterations in both DM1-derived fibroblasts and normal fibroblasts transiently expressing CUG repeats.
Materials and methods
Cell culture and transfection
Control [GM02673 (CTG5/19)] and DM1-derived skin fibroblasts [GM04033 (CTG1000) and GM03132 (CTG2000)] were purchased from Coriell Cell Repositories (Camden, NJ, USA). The length of the repeats in cell cultures was confirmed by Triplet repeat primed PCR as previously [23]. Fibroblast cultures were maintained in minimal essential medium (MEM; Invitrogen, Carlsbad, CA, USA) supplemented with 15 % fetal bovine serum and nonessential amino acids at 37 °C in a humidified 5 % CO2 atmosphere. For transfection, primary fibroblasts were seeded onto glass coverslips and grown overnight to ~50 % confluence. On the following day, fibroblasts were transfected with 2 µg of DNA pre-mixed with 4 µl of lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) in 100 µl serum-free MEM medium for 20 min at room temperature. After 1 h, the cells were washed and changed to complete MEM medium for 48 h prior to analysis.
RNA extraction and quantitative polymerase chain reaction (RT-qPCR) analysis
Total RNA was isolated from fibroblast cultures using TRIzol Reagent (Invitrogen, Thermo Fisher Scientific Inc. MA. USA), quantified on a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific Inc. MA. USA) and reverse transcribed using random hexanucleotides and the M-MLV reverse transcriptase (Invitrogen), according to manufacturer’s instructions. PCR was carried out in 25 µl, containing 12.5 μl Maxima SYBR Green/ROX qPCR Master Mix 2X, 1.5 μl cDNA, and 100 nM of each primer in a StepOnePlus™ Real-Time PCR System (Applied Biosystems). Expression of lamins A and C was quantified by the 2ΔΔct method normalized to GAPDH. Primer sequences were as follow: lamin A, forward 5′-CACTGGGGAAGAAGTGGCCA-3′ and reverse 5′-GAGCCGTGGTGGTGATGGAG-3′; lamin C forward 5′-CACTGGGGAAGAAGTGGCCA-3′ and reverse 5′ TACCACTCACGTGGTGGTGA-3′ and GAPDH, forward 5′-CGCTCTCTGCTCCTCCTGTT-3′ and reverse 5′-CCATGGTGTCTGAGCGATGT-3′.
Antibodies and vectors
The following primary antibodies were used: rabbit polyclonal antibodies anti-lamin A/C (H-110; 1:4,000 for WB), anti-emerin (FL-254; 1:25 for IF) and anti-lamin B1 (H90; 1:25 for IF), as well as a mouse polyclonal anti-GAPDH (6C5; 1:2,000 for WB) were purchased from Santa Cruz Biotechnology (Santa Cruz Biotechnology, Inc., CA, USA). Rabbit polyclonal anti-fibrillarin (ab-5821 1:50 for IF) and mouse monoclonal anti-lamin A/C (1:250 dilution for IF) antibodies were acquired from Abcam (Abcam, Cambridge, MA, USA), while mouse monoclonal anti-actin antibody [24] (1:2,000 for WB) was kindly provided by Dr. Manuel Hernández from CINVESTAV.
Vectors pQBI960 and pQBIDM, expressing the DMPK 3′ UTR (exons 13–15) with 960 CTG or with no CTG repeats respectively, were generated by subcloning a BamHI–BamHI (0 CTG repeats) or BamHI–HindIII (960 CTG repeats) restriction fragment into pDGFP [25], using DT960 and DMPKS plasmids [26] as templates respectively.
Immunofluorescence and confocal microscopy analysis
Cells grown on coverslips were fixed with 4 % paraformaldehyde for 10 min and permeabilized by exposure to 0.2 % Triton X-100 in PBS for 10 min at room temperature. Cells were then incubated overnight at 4 °C with the appropriate primary antibody and the following day washed with PBS and incubated for 1 h at room temperature with a fluorescein-conjugated goat anti-mouse or -rabbit IgG (Zymed Laboratories, Inc. San Francisco, CA, USA). Nuclei were counterstained with 0.2 μg/μl DAPI (Sigma-Aldrich, St Louis, Missouri, USA) for 7 min at room temperature and washed with PBS, and then cell preparations were mounted on microscope slides with VectaShield (Vector Laboratories, Inc., Burlingame, CA, USA) prior to being examined on a confocal laser scanning microscope (TCP-SP5 or TCS-SP8 Leica, Heidelberg, Germany) employing a Plan Neo Fluor 63× (NA = 1.4) oil-immersion objective.
For nuclear envelope morphological analysis, 500 cells from each culture (control and DM1-derived fibroblasts) were analyzed. Fibroblast nuclei were scored as normal if showed the typical ring-like immunostaining pattern for lamin A/C, lamin B1 or emerin, and their appearance under DAPI-staining was spheroid or ellipsoid. On the other hand, nuclei were scored abnormal if exhibited intense nucleoplasmic staining, invaginations, lobules, and/or blebs under lamin A/C, lamin B1 or emerin immunolabeling analysis; and/or if they were at least twice larger than those in control cells under DAPI staining examination. Likewise, 500 cells from each culture (control and DM1-derived fibroblasts) were scored for nucleoli morphological analysis. Fibroblast nucleoli were scored as normal if 2–3 large rounded nucleoli were visible per cell after fibrillarin-immunostaining, and scored as distorted if cells contain a single elongated nucleolus or numerous dispersed small nucleoli after fibrillarin-immunostaining, as previously [27].
FISH (fluorescence in situ hybridization) and FISH-IF
The cells grown on coverslips were fixed with 4 % paraformaldehyde for 10 min and permeabilized by exposure to cold 2 % acetone in PBS for 5 min. Cell preparations were incubated in pre-hibrydation buffer (SSC 2×, 30 % formamide) for 10 min at room temperature and then incubated with hibrydation buffer [SSC 2×, 30 % formamide, 0.02 % BSA, 2 mM vanadyl ribonucleoside (Sigma-Aldrich Inc., MO, USA), 66 μg/ml yeast tRNA (Sigma-Aldrich Inc.), and 1 ng/μl Cy3-conjugated CAG6 probe (Sigma-Aldrich Inc.)] for 2 h in a humidified chamber at 37 °C. Preparations were washed as follows: twice in prehybridation buffer at 42 °C, twice in SCC 1× at room temperature, and once in PBS. For immunofluorescence after FISH, the slides were incubated in PBS with 0.5 % gelatin and 1.5 % BSA for 20 min at room temperature and then incubated overnight at 4 °C with primary anti-lamin A/C antibody. After washing three times with PBS cell preparations were incubated for 1 h at room temperature with a fluorescein-conjugated anti-mouse IgG (Zymed Laboratories, Inc., San Francisco, CA, USA). Nuclei were counterstained with DAPI (0.2 μg/μl, Sigma-Aldrich Inc.) for 7 min at room temperature, washed with PBS, and mounted on microscope slides with VectaShield (Vector Laboratories, Inc., Burlingame, CA, USA) for confocal microscopy analysis.
Western blotting
The cells were centrifuged and resuspended in 50 μl of lysis buffer (10 mM Tris-HCl pH 8.0, 2 mM MgCl2, 1 % Triton ×100, 1× Complete and 1 mM PMSF protease inhibitors cocktail). Homogenates were clarified by centrifugation at 15,000×g for 2 min at 4 °C, and protein concentrations were determined by the Bradford method. Protein extracts were electrophoresed on 10 % SDS–polyacrylamide gels and transferred onto nitrocellulose membranes (BIO-RAD, Germany). Membranes were blocked in TBST [100 mM Tris–HCl pH 8.0, 150 mM NaCl, 0.5 % (v/v) Tween-20] with 10 % (w/v) low-fat dried milk and then incubated overnight at 4 °C with the appropriate primary antibody, and the following day washed three times with TBST and incubated for 1 h at room temperature with the appropriate secondary antibody and then washed three times with TBST. Specific proteins were visualized using the enhanced chemiluminescence (ECL™) western blotting detection system (Amersham Pharmacia, GE Healthcare) according to the manufacturer’s instructions.
Morphometric analysis
To analyze changes in nuclear shape, 500 randomly selected nuclei from each culture were measured as previously [28–30]. The nuclear contour ratio (4π × area/perimeter2) and the crossed diameter ratio (length/width) were calculated using the measure function in the NIS-Elements AR Version 4.13.01 (Nikon) Software. Objects were selected automatically using the smart threshold function and reviewed manually using the binary menu functions.
Statistical analysis
Statistical analyses were performed using the two-tailed Mann–Whitney test and the GraphPad Prism 6 software (La Jolla California USA, www.graphpad.com), with the exception of quantitative reverse transcription PCR (qRT-PCR) assays that were analyzed using one-way analysis of variance (one-way ANOVA).
Results
Altered nuclear morphology in DM1-derived fibroblasts
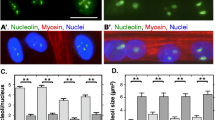
To examine whether DM1 gene mutation impacts nuclear morphology, fibroblasts derived from patients harboring mutant alleles with CTG1000 or CTG2000 repeats were immunolabeled for lamin A/C, and further analyzed by confocal microscopy, using fibroblast from age-matched healthy subject containing alleles with 5 and 19 CTG repeats (CTG5/19) as control. Confocal imaging revealed increased number of DM1-derived fibroblast nuclei showing irregular immunolabeling pattern for lamin A/C, characterized by increased immunostaining at both NE and nucleoplasm, invaginations of the NE, and an apparent augmented size of nuclei, compared with control fibroblasts (Fig. 1a). Quantitative analysis showed 30.8 and 23.7 % of nuclei with altered localization of lamin A/C in DM1-derived fibroblasts with CTG1000 and CTG2000 repeats respectively, compared to only about 12.1 % in control fibroblasts (Fig. 1b). Irregular NE immunostaining in DM1-derived fibroblasts were also evident using antibodies against lamin B1 and emerin, including increased immunolabeling and NE invaginations (Fig. 2a).
Altered localization of lamin A/C in DM1-derived fibroblasts. a Control (CTG5/19), CTG1000 and CTG2000 DM1 fibroblasts were seeded on coverslips and subjected to immunofluorescence analysis using anti-lamin A/C antibodies. Cells were counterstained with DAPI for nuclei visualization prior to being analyzed by confocal microscopy. All images were collected using identical acquisition settings. Representative single typical optical Z-sections were selected to show distribution of lamin A/C. Scale bar 10 μm. b The percentage of cells with abnormal staining of lamin A/C is shown. Results represent the mean ± SD for three separate experiments (n = 500 cells), with significant differences between control and DM1-derived fibroblasts denoted by the p value (two-tailed Mann–Whitney test). c Lysates from control (CTG5/19), CTG1000 and CTG2000 DM1 fibroblasts were analyzed by SDS-PAGE/western blot using specific antibodies against lamin A/C. d Graph showing lamin A/lamin C expression ratio at mRNA and protein levels in control and either CTG1000 and CTG2000 DM1-derived fibroblasts. Messenger RNA expression of lamin A and lamin C was examined by qRT-PCR using GAPDH as control (see “Materials and methods” section), while lamin A/lamin C protein levels were calculated by densitometric analysis of immunoblots shown in (c), using GAPDH as loading control. Results represent the mean ± SEM of three separate experiments, with relative expression obtained in control cells set at 1. No significant differences between control and DM1-derived fibroblasts were determined by one-way ANOVA
Lamin B1 and emerin exhibited altered distribution in DM1-derived fibroblasts. a Control (CTG5/19) and CTG2000 DM1-derived fibroblasts were seeded on coverslips, and subjected to immunofluorescence analysis using specific antibodies against lamin B1 or emerin. Cells were counterstained with DAPI for nuclei visualization prior to being analyzed by confocal microscopy. All images were collected using identical acquisition settings, and single typical optical Z-sections were selected to show distribution of lamin B1 and emerin. Scale bar, 10 μm. b The percentage of cells with abnormal staining of lamin B1 and emerin is shown. Results represent the mean ± SD for three separate experiments (n = 500 cells), with significant differences between control and DM1-derived fibroblasts denoted by the p values (two-tailed Mann–Whitney test)
A quantitative analysis showed significantly higher percentage of cells with altered localization of both lamin B1 and emerin in CTG2000 DM1 than in control cultures (Fig. 2b) Morphometric analysis of 500 randomly chosen nuclei revealed a subtle but statistically significant differences in crossed diameter and nuclear contour ratios between control and DM1-derived fibroblasts (Table 1); however, selective analysis of 30 DM1 nuclei that exhibited altered localization of lamin A/C revealed greater alterations in crossed diameter (2.00 ± 0.03 and 2.46 ± 0.1 for control and CTG2000 DM1 fibroblasts respectively, p = < 0.0001) and nuclear contour (0.94 ± 0.005 and 0.86 ± 0.01 for control and CTG2000 DM1 fibroblasts respectively, p = < 0.0001) ratios. Increased crossed diameter ratio reflects an augment in size, while decreased contour ratio indicates nuclear deformation (i.e. blebbed or lobed nuclei) [28–30]. Overall these data imply that DM1 mutation affects global NE organization and ultimately nuclear morphology. In spite of the increased immunolabeling and mislocalization of lamin A/C found in DM1-derived fibroblasts, both qRT-PCR and western blotting assays showed similar lamin A/lamin C expression ratio between control and either CTG1000 and CTG2000 DM1 fibroblasts (Fig. 1c, d).
Nuclear foci containing CUG expanded repeats induce nuclear morphology defects
To ascertain whether nuclear shape abnormalities are related with expression of expanded CUG repeats, in situ hybridization assay using a fluorescent-labeled (CAG)6 probe (FISH), follow by immunofluorescence assay (FISH-IF) for lamin A/C was carried out in CTG2000 DMI fibroblasts. Interestingly, the presence of nuclear foci appears to be directly related with nuclear shape abnormalities (Fig. 3a); the higher the number of nuclear foci, the higher the percentage of misshapen nuclei (Fig. 3b). To test directly the hypothesis that expression of CUG expanded repeats induce nuclei morphology defects, control fibroblasts (CTG5/19) were transiently transfected with vector expressing the DMPK 3′ UTR containing 960 interrupted CTG repeats, and further subjected to FISH-IF. Transient expression of CTG-expanded repeats resulted in altered distribution of lamin A/C and abnormal nuclear shape, while typical ring-like distribution of lamin A/C in the NE and the presence of oval shaped nuclei were consistently observed in control fibroblasts transiently-transfected with vector expressing the DMPK 3′ UTR containing no CTG repeats (Fig. 4). A quantitative analysis showed 37.00 ± 2.7 and 79.27 ± 5.2 % of misshapen nuclei in control and DM1-derived fibroblasts respectively (right panel).
Presence of nuclear foci is related with mislocalization of lamin A/C and nuclear shape defects. a Immunolocalization of lamin A/C and in situ hybridization of CUG repeats using a (CAG)6 probe were carried out in single nuclei of CTG2000 DM1 fibroblasts. Cells were counterstained with DAPI for nuclei visualization prior to being analyzed by confocal microscopy. All images were collected using identical acquisition settings, and representative single typical optical Z-sections were selected to show the presence of nuclear foci (1–4 or >5, arrows) and distribution of lamin A/C in the same nucleus. b The percentage of cells with abnormal staining of lamin A/C with respect to the number of nuclear foci per nucleus (1-4 or > 5) is shown. Results represent the mean ± SD for 3 separate experiments (n = 100 cells), with significant differences denoted by the p value (two-tailed Mann–Whitney test)
Transient expression of expanded CUG repeats induces lamin A/C mislocalization and altered nuclear morphology. a Control fibroblasts seeded on coverslips were transfected with vector expressing the DMPK 3′ UTR containing ~960 interrupted CTG repeats (CTG960) or lacking CTG repeats (CTG0). Immunolabeling for lamin A/C and in situ hybridization of CUG repeats using a (CAG)6 probe were carried out in single nuclei. Cells were stained with DAPI to enable nuclei visualization prior to being analyzed by confocal microscopy. All images were collected using identical acquisition settings. Typical optical Z-sections showing lamin A/C distribution and nuclear foci in the same nucleus are shown. Scale bar 10 μm. b The percentage of fibroblasts with abnormal staining of lamin A/C is shown (n = 100 cells), with significant differences denoted by the p value (two-tailed Mann–Whitney test)
Nucleoli disruption in DM1-derived fibroblasts
To ascertain whether CTG-expanded repeats alter the organization of other subnuclear structures, the structure of nucleoli was analyzed by immunostaining nucleoli in control and CTG2000 DM1 fibroblasts using antibodies against the nucleolar protein fibrillarin. DM1-derived fibroblasts exhibited high percentage of distorted nucleoli, compared with control fibroblasts (Fig. 5; 36.0 ± 2.17 and 55.77 ± 2.08 % for control and DM1 fibroblasts respectively), which implies that expanded CTG repeats altered the spatial distribution of nucleoli.
Spatial organization of nucleoli is disrupted in DM1-derived fibroblasts. a Control (CTG5/19) and CTG2000 DM1 fibroblasts grown on coverslips were subjected to immunofluorescence/confocal microscopy analysis using specific antibodies against fibrillarin. All images were collected using identical acquisition settings and representative optical Z-sections showing fibrillarin localization are shown. Scale bar 10 μm. b The percentage of cells containing distorted nucleoli is shown. Results represent the mean ± SD for three separate experiments (n = 500 cells), with significant differences between control and CTG2000 fibroblasts denoted by the p value (two-tailed Mann–Whitney test)
Discussion
In this study we provide for the first time evidence showing both aberrant nuclear morphology and disorganization of nucleoli in DM1 cells. We found altered NE organization in a fraction of 30 % of fibroblasts derived from DM1 patients, evidenced by increased immunolabeling and mislocalization of the NE proteins, lamins A/C and B1 and emerin. Although metric measurements showed subtle changes in nuclear shape in DM1-derived cells, greater nuclear morphology alterations that resemble those reported in laminopathies [28–30] were found in the fraction of DM1 nuclei that displayed mislocalization of lamin A/C. Interestingly, the occurrence of misshapen nuclei was more frequent in DM1-fibroblasts containing numerous nuclear foci, which implicates the mutant DMPK mRNA in the development of nuclear morphology defects. A recent work showed that depletion of DPMK result in NE disruption, implicating the DM1 gene protein in NE stability [31]. Thus, it is possible that haploinsufficiency of DMPK in DM1-derived fibroblasts might alter NE organization. Nevertheless, that NE disorganization was induced in control fibroblasts through transient overexpression of the DMPK 3′ UTR containing 960 CTG but not with the 3′ UTR lacking CTG repeats implies that expression of the toxic expanded mRNA rather than reduction of DMPK is critical for NE impairing in our cell model.
Although molecular mechanisms that participate in defining nuclear shape have remained unclear, the important role of NE components in determining nuclear architecture is highlighted by the abnormal shaped nuclei observed in a variety of human disease, collectively called laminopathies, where genes encoding lamin or lamin-binding proteins are mutated [32, 33]. It is difficult at this point to hypothesize how the expression of CUG-containing mutant mRNA impacts nuclear morphology; however, in the light of the current knowledge of molecular bases underlying DM1 it could be speculated that aberrant RNA splicing caused by the expanded RNA [7] might affect the alternative splicing of LMNA gene, resulting in a shift in the balance of lamin A/C ratio, and ultimately in nuclear lamin disorganization. Nevertheless, qRT-PCR and immunoblotting analyses showing similar lamin A/lamin C expression ratio at mRNA and protein level between control and DM1-derived fibroblasts is inconsistent with this possibility. As lipins, a family of lipid phosphatases, have been implicated in the maintenance of nuclear shape [34–36], it should be noted that aberrant alternative splicing/gene expression of genes involved in lipid metabolism was found in a mouse model of DM1, including LPIN1, ATP9a, PICALM, and ACSL5 genes [37]. Clearly, the demonstration that the occurrence of mis-splicing events in genes involved in lipid metabolism in DM1 fibroblasts and its potential connection with nuclear membrane morphology requires further investigation.
An alternative likely mechanism to explain impaired nuclear integrity in DM1 fibroblasts is the effect exerted by the expanded RNA on chromatin dynamics, including altered nucleosome positioning [38–40] and chromatin remodeling [21]. Experimental evidence showing that conformational changes in chromatin structure are at least partially responsible for shaping the nucleus has recently been accumulated: downregulation of either BAF, an adaptor protein between nuclear lamin and chromatin, and p55, a component of histone modifying and chromatin assembly complexes, alters nuclear morphology in Drosophila cells [41]. Likewise, decreased expression of the chromatin remodeling ATPase BRG1 causes nuclear shaped changes in MCF-10A cells [42]. Furthermore, overexpression of Esc1p, an anchor for chromatin results in abnormal nuclear morphology in yeast cells [43]. Additional experiments focusing on setting out alterations in chromatin structure in DM1 fibroblasts and their potential connection with nuclear morphology defects will be undertaken in our laboratory.
Altered lamina A/C distribution in DM1 fibroblasts with a direct effect on nuclear structure implies that DM1 pathogenesis might share pathological mechanisms with muscular dystrophies caused by mutations in the LMNA gene, including different Emery-Dreifuss muscular dystrophies and limb-girdle muscular dystrophy type 1B. Besides some clinical features that are shared between DM1 and skeletal muscle laminopathies, including muscle weakness and wasting and cardiomyopathy [44, 45], it appears that altered myogenesis signaling pathway is a common molecular mechanism to the two types of muscular dystrophy. On the one hand, it has been shown that overexpression of the mutant DMPK 3′UTR mRNA disrupts myoblast fusion in both myoblast cell line C2C12 [46] and primary myoblasts from transgenic mice [47]. At molecular level, expression of the mutant DMPK 3′UTR mRNA results in downregulation of both the cell cycle regulator factor p21 [48] and the key myogenic transcription factor MyoD [49], which ultimately impair muscle differentiation. On the other hand, it is thought that mutation in the LMNA gene disrupts the interaction of lamin A/C and its partner LAP2α with different transcription factors and chromatin modifying complexes, leading to altered regulation of gene expression during muscle differentiation (reviewed in [50]). Supporting this idea, myoblasts generated from Lmna −/− mice exhibit decreased levels of proteins necessary for muscle cell differentiation, including MyoD, desmin, pRB and M-cadherin [51].
Fibroblasts derived from DM1 patients also exhibit nucleoli disorganization. The nucleolus is a multifunctional nuclear body with multiple roles, including ribosome biogenesis, cell proliferation, cell growth and survival, and the response to stress [52]. Importantly, disruption of nucleolar integrity is associated with aging and early response to cellular stress, and is underlying several human diseases, including neurodegenerative disorders and cancer [53]. Although the impact of the DMPK gene expanded RNA in specific functions associated with nucleoli remains to be elucidated, it is possible that mislocalization of lamin B1 observed in DM1 fibroblasts causes in turn altered nucleoli structure, because lamin B1 is involved in nucleoli organization [54], preserving its functional plasticity [27]. Interestingly, expanded mutant Huntingtin transcript interacts with the nucleolar protein nucleolin and this aberrant interaction leads to down-regulation of pre-45s rRNA transcription [55]. Thus, the possibility that the DMPK mutant mRNA sequesters components of nucleoli altering their architecture can not be ruled out.
In conclusion, we describe to the best of our knowledge for the first time alterations in the organization of the nuclear envelope and nucleoli in DM1 cells, and demonstrate that expression of the expanded DMPK gene RNA is underlying nuclear structure alterations. It is thought that impaired NE organization results in altered DNA repair, gene expression, and cell cycle, and ultimately causes apoptosis and aneuploidy [32, 33] Therefore, definition of the physiological relevance of altered NE disorganization in DM1 requires further investigation of NE-dependent processes in DM1 models.
References
Harper PS, van Engelen BG, Eymard B, Rogers M, Wilcox D (2002) In: 99th ENMC international workshop: myotonic dystrophy: present management, future therapy. 9–11 November 2001, Naarden. Neuromuscular disorders: NMD 12(6):596–599
Schara U, Schoser BG (2006) Myotonic dystrophies type 1 and 2: a summary on current aspects. Semin Pediatr Neurol 13(2):71–79. doi:10.1016/j.spen.2006.06.002 S1071-9091(06)00092-1 [pii]
Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, Hudson T et al (1992) Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell 68(4):799–808 0092-8674(92)90154-5 [pii]
Tian B, White RJ, Xia T, Welle S, Turner DH, Mathews MB, Thornton CA (2000) Expanded CUG repeat RNAs form hairpins that activate the double-stranded RNA-dependent protein kinase PKR. RNA 6(1):79–87
Cho DH, Tapscott SJ (2007) Myotonic dystrophy: emerging mechanisms for DM1 and DM2. Biochim Biophys Acta 1772(2):195–204. doi:10.1016/j.bbadis.2006.05.013 S0925-4439(06)00098-6 [pii]
Magana JJ, Cisneros B (2011) Perspectives on gene therapy in myotonic dystrophy type 1. J Neurosci Res 89(3):275–285. doi:10.1002/jnr.22551
Sicot G, Gomes-Pereira M (2013) RNA toxicity in human disease and animal models: from the uncovering of a new mechanism to the development of promising therapies. Biochim Biophys Acta 1832(9):1390–1409. doi:10.1016/j.bbadis.2013.03.002
Fu YH, Friedman DL, Richards S, Pearlman JA, Gibbs RA, Pizzuti A, Ashizawa T, Perryman MB, Scarlato G, Fenwick RG Jr et al (1993) Decreased expression of myotonin-protein kinase messenger RNA and protein in adult form of myotonic dystrophy. Science 260(5105):235–238
Reddy S, Smith DB, Rich MM, Leferovich JM, Reilly P, Davis BM, Tran K, Rayburn H, Bronson R, Cros D, Balice-Gordon RJ, Housman D (1996) Mice lacking the myotonic dystrophy protein kinase develop a late onset progressive myopathy. Nat Genet 13(3):325–335. doi:10.1038/ng0796-325
Berul CI, Maguire CT, Gehrmann J, Reddy S (2000) Progressive atrioventricular conduction block in a mouse myotonic dystrophy model. J Interv Card Electrophysiol Int j Arrhythm Pacing 4(2):351–358
Ho TH, Bundman D, Armstrong DL, Cooper TA (2005) Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum Mol Genet 14(11):1539–1547. doi:10.1093/hmg/ddi162
Philips AV, Timchenko LT, Cooper TA (1998) Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science 280(5364):737–741
Savkur RS, Philips AV, Cooper TA (2001) Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat Genet 29(1):40–47. doi:10.1038/ng704
Savkur RS, Philips AV, Cooper TA, Dalton JC, Moseley ML, Ranum LP, Day JW (2004) Insulin receptor splicing alteration in myotonic dystrophy type 2. Am J Hum Genet 74(6):1309–1313. doi:10.1086/421528
Timchenko NA, Cai ZJ, Welm AL, Reddy S, Ashizawa T, Timchenko LT (2001) RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J Biol Chem 276(11):7820–7826. doi:10.1074/jbc.M005960200
Carpentier C, Ghanem D, Fernandez-Gomez FJ, Jumeau F, Philippe JV, Freyermuth F, Labudeck A, Eddarkaoui S, Dhaenens CM, Holt I, Behm-Ansmant I, Marmier-Gourrier N, Branlant C, Charlet-Berguerand N, Marie J, Schraen-Maschke S, Buee L, Sergeant N, Caillet-Boudin ML (2014) Tau exon 2 responsive elements deregulated in myotonic dystrophy type I are proximal to exon 2 and synergistically regulated by MBNL1 and MBNL2. Biochim Biophys Acta 1842(4):654–664. doi:10.1016/j.bbadis.2014.01.004
Echeverria GV, Cooper TA (2014) Muscleblind-like 1 activates insulin receptor exon 11 inclusion by enhancing U2AF65 binding and splicing of the upstream intron. Nucleic Acids Res 42(3):1893–1903. doi:10.1093/nar/gkt1020
Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS, Brook JD (2002) Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum Mol Genet 11(7):805–814
Ebralidze A, Wang Y, Petkova V, Ebralidse K, Junghans RP (2004) RNA leaching of transcription factors disrupts transcription in myotonic dystrophy. Science 303(5656):383–387
Mankodi A, Urbinati CR, Yuan QP, Moxley RT, Sansone V, Krym M, Henderson D, Schalling M, Swanson MS, Thornton CA (2001) Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum Mol Genet 10(19):2165–2170
Brouwer JR, Huguet A, Nicole A, Munnich A, Gourdon G (2013) Transcriptionally Repressive Chromatin Remodelling and CpG Methylation in the Presence of Expanded CTG-Repeats at the DM1 Locus. J nucleic acids 2013:567435. doi:10.1155/2013/567435
Schneider R, Grosschedl R (2007) Dynamics and interplay of nuclear architecture, genome organization, and gene expression. Genes Dev 21(23):3027–3043. doi:10.1101/gad.1604607
Magana JJ, Cortes-Reynosa P, Escobar-Cedillo R, Gomez R, Leyva-Garcia N, Cisneros B (2011) Distribution of CTG repeats at the DMPK gene in myotonic dystrophy patients and healthy individuals from the Mexican population. Mol Biol Rep 38(2):1341–1346. doi:10.1007/s11033-010-0235-7
Garcia-Tovar CG, Perez A, Luna J, Mena R, Osorio B, Aleman V, Mondragon R, Mornet D, Rendon A, Hernandez JM (2001) Biochemical and histochemical analysis of 71 kDa dystrophin isoform (Dp71f) in rat brain. Acta Histochem 103(2):209–224
Acosta R, Montanez C, Fuentes-Mera L, Gonzalez E, Gomez P, Quintero-Mora L, Mornet D, Alvarez-Salas LM, Cisneros B (2004) Dystrophin Dp71 is required for neurite outgrowth in PC12 cells. Exp Cell Res 296(2):265–275. doi:10.1016/j.yexcr.2004.01.015
Ho TH, Savkur RS, Poulos MG, Mancini MA, Swanson MS, Cooper TA (2005) Colocalization of muscleblind with RNA foci is separable from mis-regulation of alternative splicing in myotonic dystrophy. J Cell Sci 118(Pt 13):2923–2933
Martin C, Chen S, Maya-Mendoza A, Lovric J, Sims PF, Jackson DA (2009) Lamin B1 maintains the functional plasticity of nucleoli. J Cell Sci 122(Pt 10):1551–1562. doi:10.1242/jcs.046284
Lammerding J, Hsiao J, Schulze PC, Kozlov S, Stewart CL, Lee RT (2005) Abnormal nuclear shape and impaired mechanotransduction in emerin-deficient cells. J cell biol 170(5):781–791. doi:10.1083/jcb.200502148
Cui Y, Koop EA, van Diest PJ, Kandel RA, Rohan TE (2007) Nuclear morphometric features in benign breast tissue and risk of subsequent breast cancer. Breast Cancer Res Treat 104(1):103–107. doi:10.1007/s10549-006-9396-4
Broers JL, Peeters EA, Kuijpers HJ, Endert J, Bouten CV, Oomens CW, Baaijens FP, Ramaekers FC (2004) Decreased mechanical stiffness in LMNA−/− cells is caused by defective nucleo-cytoskeletal integrity: implications for the development of laminopathies. Hum Mol Genet 13(21):2567–2580. doi:10.1093/hmg/ddh295
Harmon EB, Harmon ML, Larsen TD, Yang J, Glasford JW, Perryman MB (2011) Myotonic dystrophy protein kinase is critical for nuclear envelope integrity. J Biol Chem 286(46):40296–40306. doi:10.1074/jbc.M111.241455
Chi YH, Chen ZJ, Jeang KT (2009) The nuclear envelopathies and human diseases. J Biomed Sci 16:96. doi:10.1186/1423-0127-16-96
Schreiber KH, Kennedy BK (2013) When lamins go bad: nuclear structure and disease. Cell 152(6):1365–1375. doi:10.1016/j.cell.2013.02.015
Siniossoglou S (2009) Lipins, lipids and nuclear envelope structure. Traffic 10(9):1181–1187. doi:10.1111/j.1600-0854.2009.00923.x
Santos-Rosa H, Leung J, Grimsey N, Peak-Chew S, Siniossoglou S (2005) The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J 24(11):1931–1941. doi:10.1038/sj.emboj.7600672
Golden A, Liu J, Cohen-Fix O (2009) Inactivation of the C. elegans lipin homolog leads to ER disorganization and to defects in the breakdown and reassembly of the nuclear envelope. J Cell Sci 122(Pt 12):1970–1978. doi:10.1242/jcs.044743
Du H, Cline MS, Osborne RJ, Tuttle DL, Clark TA, Donohue JP, Hall MP, Shiue L, Swanson MS, Thornton CA, Ares M Jr (2010) Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol 17(2):187–193
Tomita N, Fujita R, Kurihara D, Shindo H, Wells RD, Shimizu M (2002) Effects of triplet repeat sequences on nucleosome positioning and gene expression in yeast minichromosomes. Nucleic Acids Res Suppl 2:231–232
Wang YH, Amirhaeri S, Kang S, Wells RD, Griffith JD (1994) Preferential nucleosome assembly at DNA triplet repeats from the myotonic dystrophy gene. Science 265(5172):669–671
Wang YH, Griffith J (1995) Expanded CTG triplet blocks from the myotonic dystrophy gene create the strongest known natural nucleosome positioning elements. Genomics 25(2):570–573
Polychronidou M, Hellwig A, Grosshans J (2010) Farnesylated nuclear proteins kugelkern and lamin Dm0 affect nuclear morphology by directly interacting with the nuclear membrane. Mol Biol Cell 21(19):3409–3420. doi:10.1091/mbc.E10-03-0230
Imbalzano KM, Cohet N, Wu Q, Underwood JM, Imbalzano AN, Nickerson JA (2013) Nuclear shape changes are induced by knockdown of the SWI/SNF ATPase BRG1 and are independent of cytoskeletal connections. PLoS One 8(2):e55628. doi:10.1371/journal.pone.0055628
Taddei A, Hediger F, Neumann FR, Bauer C, Gasser SM (2004) Separation of silencing from perinuclear anchoring functions in yeast Ku80, Sir4 and Esc1 proteins. EMBO J 23(6):1301–1312. doi:10.1038/sj.emboj.7600144
Harper PS (1989) Myotonic dystrophy. 2nd edn. W. B. Saunders (pub.), Philadelphia
Worman HJ, Bonne G (2007) “Laminopathies”: a wide spectrum of human diseases. Exp Cell Res 313(10):2121–2133. doi:10.1016/j.yexcr.2007.03.028
Amack JD, Paguio AP, Mahadevan MS (1999) Cis and trans effects of the myotonic dystrophy (DM) mutation in a cell culture model. Hum Mol Genet 8(11):1975–1984
Storbeck CJ, Drmanic S, Daniel K, Waring JD, Jirik FR, Parry DJ, Ahmed N, Sabourin LA, Ikeda JE, Korneluk RG (2004) Inhibition of myogenesis in transgenic mice expressing the human DMPK 3′-UTR. Hum Mol Genet 13(6):589–600. doi:10.1093/hmg/ddh064
Timchenko NA, Iakova P, Cai ZJ, Smith JR, Timchenko LT (2001) Molecular basis for impaired muscle differentiation in myotonic dystrophy. Mol Cell Biol 21(20):6927–6938
Amack JD, Reagan SR, Mahadevan MS (2002) Mutant DMPK 3′-UTR transcripts disrupt C2C12 myogenic differentiation by compromising MyoD. Journal Cell Biol 159(3):419–429. doi:10.1083/jcb.200206020
Dubinska-Magiera M, Zaremba-Czogalla M, Rzepecki R (2013) Muscle development, regeneration and laminopathies: how lamins or lamina-associated proteins can contribute to muscle development, regeneration and disease. Cell Mol Life Sci CMLS 70(15):2713–2741. doi:10.1007/s00018-012-1190-3
Frock RL, Kudlow BA, Evans AM, Jameson SA, Hauschka SD, Kennedy BK (2006) Lamin A/C and emerin are critical for skeletal muscle satellite cell differentiation. Genes Dev 20(4):486–500. doi:10.1101/gad.1364906
Morimoto M, Boerkoel CF (2013) The role of nuclear bodies in gene expression and disease. Biology 2(3):976–1033. doi:10.3390/biology2030976
Quin JE, Devlin JR, Cameron D, Hannan KM, Pearson RB, Hannan RD (2014) Targeting the nucleolus for cancer intervention. Biochim Biophys Acta. doi:10.1016/j.bbadis.2013.12.009
Tang CW, Maya-Mendoza A, Martin C, Zeng K, Chen S, Feret D, Wilson SA, Jackson DA (2008) The integrity of a lamin-B1-dependent nucleoskeleton is a fundamental determinant of RNA synthesis in human cells. J Cell Sci 121(Pt 7):1014–1024. doi:10.1242/jcs.020982
Tsoi H, Lau TC, Tsang SY, Lau KF, Chan HY (2012) CAG expansion induces nucleolar stress in polyglutamine diseases. Proc Natl Acad Sci USA 109(33):13428–13433. doi:10.1073/pnas.1204089109
Acknowledgments
This work was supported by Science and Technology Institute of Mexico City Grant (PIFUTP08-164). We thank Dr. Thomas Cooper (Baylor College of Medicine) for donation of DMPKS and DT960 vectors.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rodríguez, R., Hernández-Hernández, O., Magaña, J. et al. Altered nuclear structure in myotonic dystrophy type 1-derived fibroblasts. Mol Biol Rep 42, 479–488 (2015). https://doi.org/10.1007/s11033-014-3791-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3791-4