Abstract
Phosphatidylinositol 3-kinase (PI3K) plays an important role in the metabolic actions of insulin and is required for adipogenesis. Regulatory subunit 1 of PI3K (PIK3R1) is a critical component of the PI3K signaling pathway. Peroxisome proliferator-activated receptor gamma (PPARγ) is a key regulator of adipogenesis. Although the PPARγ agonist rosiglitazone induces the expression of PIK3R1, the transcriptional regulation of PIK3R1 in adipocytes remains unknown. In this study, we investigated whether PIK3R1 is a direct target of PPARγ. The level of PIK3R1 expression in 3T3-L1 cells was increased after the induction of adipocyte differentiation and was also induced by overexpression of PPARγ. Furthermore, the upregulation of PPARγ-mediated PIK3R1 expression enhanced the insulin-stimulated AKT activation in 3T3-L1 cells. Two putative peroxisome proliferator response elements (PPREs) in the PIK3R1 promoter were identified as PPARγ binding sites. By chromatin immunoprecipitation, we observed that PPARγ interacts with the two PPRE regions of the PIK3R1 promoter in mature adipocytes. In addition, luciferase reporter assays showed that the −1183/−1161 and −573/−551 regions of the PIK3R1 promoter contain essential elements for PPARγ binding. Taken together, these results suggest that PPARγ is essential for the transcriptional activity of PIK3R1 during adipogenesis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Adipose tissue plays a role as the major fat storage site including triglycerides, which is involved in the regulation of energy homeostasis [1, 2]. Understanding the molecular basis of obesity is important from both the physiological and clinical perspectives. Peroxisome proliferator-activated receptor gamma (PPARγ) is a key modulator of adipocyte differentiation, which activates lipogenic genes encoding proteins, such as perilipin, adipocyte fatty acid-binding protein (aFABP), and glucose transporter type 4 (GLUT4), that are required for the differentiation of adipocytes [3]. PPARγ regulates the transcriptional activity of these target genes after forming a complex with retinoid X receptor alpha (RXRα). The interaction regions of the target genes are called peroxisome proliferator response elements (PPREs), which consist of a direct repeat of the consensus half-site motif (AGGTCA) that is spaced by a single nucleotide.
Phosphatidylinositol 3-kinase (PI3K) is a component of a heterodimer composed of p110 (catalytic) and p85/regulatory subunit 1 of PI3K (PIK3R1) (regulatory) subunits. PI3K activation contributes to multiple tissue-specific biological processes, including GLUT4 translocation, the inhibition of lipolysis, and the upregulation of glycogen synthesis [4–6]. The insulin-PI3K signaling network plays a critical role in adipogenesis. Insulin activates interferon regulatory factor 1 (IRF1) by binding insulin receptor, which recruits PIK3R1 and leads to the transduction of insulin signaling [7–10]. The activation of PPARγ by rosiglitazone can induce the expression of PIK3R1 in human adipocytes [11]. However, the transcriptional regulation of PIK3R1 in the insulin signaling pathway is unclear. Therefore, in this study, we demonstrate the transcriptional activity of PIK3R1 by PPARγ in mature adipocytes.
Materials and methods
Cell culture
Cos7 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Hyclone, Logan, UT, USA) supplemented with 10 % fetal bovine serum (FBS, Hyclone, Logan, UT, USA). Cells were incubated at 37 °C and supplemented with 5 % CO2 in a humidified chamber. 3T3-L1 cells were grown in DMEM supplemented with 10 % bovine calf serum. 3T3-L1 preadipocytes were cultured until confluence and maintained for 48 h (designated as day 0). Two days after cell confluence, medium was changed to differentiation medium (1 mM dexamethasone, 10 μg/mL insulin, and 0.5 mM isobutylmethylxanthine in DMEM containing 10 % FBS). After 48 h, the differentiation medium was replaced with DMEM supplemented with 10 % FBS and 1 μg/mL insulin. Dexamethasone, insulin and isobutylmethylxanthine were purchased from Sigma (St. Louis, MO, USA).
Quantitative polymerase chain reaction (PCR)
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The cDNA was synthesized by using MMLV-reverse transcriptase (Invitrogen, Carlsbad, CA, USA) and oligo-dT primers (Promega, Madison, WI, USA). Real-time PCR quantitation was performed using the Bio-Rad CFX96 system (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s protocol. The threshold value of fluorescence was measured using iQ SYBR Green Supermix (Bio-Rad, Hercules, CA, USA). By the comparative cycle threshold method, data were proceed and presented as fold increase relative to the basal expression level. The expression level of target genes was normalized by the level of GAPDH mRNA. The following oligonucleotides were used: PIK3R1 (F) 5′-GTG CAT GGA CTG TTT CCA ATA CA-3′, (R) 5′-AAT GAC GGA CTT CTC ACT TCA C-3′; PPARγ (F) 5′-CCC TGG CAA AGC ATT TGT AT-3′, (R) 5′-TTG ATT TTA TCT TCT CCC ATC ATT-3′; GAPDH (F) 5′-GAC TTC AAC AGC AAC TCC CAC-3′, (R) 5′-TCC ACC ACC CTG TTG CTG TA-3′.
Chromatin immunoprecipitation (ChIP) assay
Preadipocytes and mature adipocytes (~107 cells) were fixed in 18.5 % formaldehyde for 10 min at room temperature and neutralized with 125 mM glycine for 5 min. After sonication, cell lysates were produced chromatin fragments averaging 500 bp in size. Fragmented samples were added into the ChIP diluted buffer [16.7 mM Tris–HCl (pH 8.1), 167 mM NaCl, 1.2 mM ethylenediaminetetraacetic acid (EDTA), 1.1 % Triton X-100, 0.01 % sodium dodecyl sulfate (SDS), and protease inhibitor cocktail]. Fragmented chromatins were incubated with anti-PPARγ antibody (Abcam, Cambridge, UK) at 4 °C. After precipitation with Protein A resin (Millipore Upstate, Lake Placid, NY, USA), immune complexes were transferred to mini columns (Bio-Rad, Hercules, CA, USA). Finally, the beads were eluted using Tris–EDTA (TE) buffer. DNA–protein cross-links were reversed by incubation with 10 % Chelex (Bio-Rad, Hercules, CA, USA) at 95 °C for 10 min. Samples were treated with proteinase K (Roche, Indianapolis, IN, USA) at 55 °C for 30 min. Precipitated chromatin was used as the template for PCR. The following oligonucleotides were used: PPRE1 (F) 5′-GTT TGC GTG TCC AAT GGG TAG-3′, (R) 5′-TGT GAG TTT TGG AAG CAG AGG G A-3′; PPRE2 (F) 5′-ACC CGG TAT GCG CCC TCC TC-3′, (R) 5′-CTT GCC TGC CCA GCT TCC CT-3′.
Luciferase reporter assay
Firefly luciferase reporter pGL3 vector was constructed to contain 1.4 k bp (pGL3-PIK3R1 1.4 kb) and 0.8 k bp (pGL3-PIK3R1 0.8 kb) from the translation start site of the mouse PIK3R1 gene. These fragments were generated by PCR using the following oligonucleotides : pGL3-PIK3R1 1.4 kb (F) 5′-GGG GTA CCT TTG CGT GTC CAA TGG GTA-3′; pGL3-PIK3R1 0.8 kb (F) 5′-GGG GTA CCG ACC TAC ATA GGC TCT GC-3′; pGL3 (R) 5′-CCA AGC TTT AGC TAC TCC TCT CCC TGA-3′. The PIK3R1 promoter (pGL3-PIK3R1 1.4 kb) was mutated by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) following the instructions provided by the supplier. Successful mutagenesis of the PIK3R1 promoter was confirmed by DNA sequencing. For the reporter assay, Cos7 cells were plated in 24 wells at a density of 5 × 104 cells per well. The reporter plasmids were transfected into Cos7 cells using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instruction. At 48 h post-transfection, luciferase activities were measured with the Dual Luciferase Assay kit (Promega, Madison, WI, USA) using a GloMax 20/20 luminometer (Turner Biosystems, Sunnyvale, CA, USA). Renilla luciferase was used for normalization.
Western blot analysis
Cells were extracted with lysis buffer (50 mM Tris–HCl, 150 mM NaCl, 1 % Nonidet P-40, 0.1 % SDS, protease inhibitor cocktail, 50 mM NaF, and 0.2 M Na3VO4). Cell lysates were separated through a 10 % SDS polyacrylamide gel and transferred onto a nitrocellulose membrane. After blocking with 5 % BSA in PBS-Tween 20, membranes were incubated with anti-PIK3R1 (1:200 dilution; GeneTex, Irvine, CA, USA), anti-phospho-AKT S473 (1:500 dilution; Cell Signaling Technology, Danvers, MA, USA), anti-AKT (1:500 dilution; Cell Signaling Technology, Danvers, MA, USA) or anti-actin (Sigma-Aldrich, St. Louis, MO, USA) antibodies in PBS-T. After washing with PBS, the samples were incubated with horseradish peroxidase—conjugated secondary antibodies. The protein signals were visualized using an enhanced chemiluminescence kit (Santa Cruz Inc., Santa Cruz, CA, USA). Densitometric analysis of western blots was performed using ImageJ program.
Statistics
Data are presented as mean ± SE of three independent experiments. The statistical significance between two groups was assessed using the Student two-tailed t test. P values less than 0.05 were considered to show a significant difference.
Results
The expression of PIK3R1 during adipocyte differentiation
In order to identify the potential regulatory actions of PIK3R1, we investigated its expression pattern in 3T3-L1 cells during adipogenesis. In preadipocytes, the signal of PIK3R1 transcript was low, whereas the expression level of PIK3R1 mRNA was significantly increased in mature 3T3-L1 cells. The level of PIK3R1 mRNA at day 6 of differentiation was increased more than fourfold compared to that in preadipocytes (Fig. 1). The level of PPARγ mRNA was also elevated by fourfold in mature adipocytes compared to that in preadipocytes. These results suggest that the expression pattern of PIK3R1 was similar to those of PPARγ during adipocyte differentiation.
The expression of PIK3R1 and PPARγ during adipogenesis. 3T3-L1 cells were stimulated to undergo adipocyte differentiation 2 days post-confluency (D0). Total RNA was prepared at the indicated time points after inducing differentiation and was subjected to quantitative real-time RT-PCR for PIK3R1 and PPARγ in triplicate. The amount of mRNA was normalized to GAPDH mRNA. Data represent the mean ± SE. *P < 0.05 versus D0. **P < 0.01 versus D0. ***P < 0.005 versus D0
Enhanced insulin-induced PIK3R1/AKT activation by PPARγ
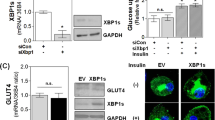
To determine the expression of PIK3R1 in cells expressing PPARγ/RXRα in 3T3-L1 cells, real-time PCR was performed. As expected, the exogenous expression of PPARγ/RXRα increased the expression of PIK3R1 mRNA in preadipocyte cells (Fig. 2a). Because PI3K is a critical component in insulin signaling and PIK3R1 is a regulatory subunit of PI3K, we next investigated the phosphorylation of AKT protein, a marker of insulin signaling, in cells expressing PPARγ/RXRα in the presence of insulin. As shown in Fig. 2b, insulin induced the phosphorylation of AKT protein in 3T3-L1 cells. The level of total AKT protein was not changed in cells regardless of PPARγ in the presence of insulin. The PPARγ/RXRα proteins did not affect the phosphorylation of AKT protein in the absence of insulin, whereas the induction of PPARγ in preadipocyte cells stimulated the insulin-induced AKT phosphorylation compared to that of empty vector (Fig. 2b). These results suggest that PPARγ stimulates the insulin signaling pathway via the induction of PIK3R1 protein as well as the phosphorylation of AKT protein.
The insulin-induced PIK3R1/AKT activation in 3T3-L1 cells overexpressing PPARγ. 3T3-L1 preadipocyte cells were transfected with empty vector (pSG) or both pSG5-PPARγ and pCMV-RXRα (PPARγ/RXRα). a Total RNA was isolated and real-time PCR was performed in triplicate. The amount of PIK3R1 mRNA was normalized to GAPDH mRNA. Data represent the mean ± SE. ***P < 0.005 versus pSG empty vector. b At 48 h post-transfection, cells were incubated in the absence or presence of insulin (100 nM) for 30 min, and then cells were lysed for SDS-PAGE. Western blot analysis was conducted with antibodies against PIK3R1, AKT, phospho-Ser473 AKT, and actin. The data are a representative example of three independent experiments. **P < 0.01 versus pSG empty vector. Ins insulin
Determination of the putative PPAR binding site in the PIK3R1 promoter
Next, we used the MatInspector program to analyze the putative PPRE sites of the PIK3R1 promoter [12]. As shown in Fig. 3a, 2 putative PPAR binding sites were identified in the 2,000 bp upstream of the PIK3R1 transcription start site: −1,183 to −1,161 (denoted as PPRE1) and −573 to −551 (denoted as PPRE2). In order to determine whether PPARγ is recruited into these PPRE sites of the PIK3R1 promoter, ChIP assay was conducted. As a result, we observed PCR products corresponding to both PPRE1 and PPRE2 in mature adipocytes but not in preadipocytes (Fig. 3b). These data suggest that PPARγ directly binds to the PPRE1 and PPRE2 regions of the PIK3R1 promoter.
Interaction of PPARγ with the putative PPRE regions in the PIK3R1 promoter. a The location of the putative PPARγ-binding sites (PPREs) is shown in the region that is −2 kb of the PIK3R1 promoter. b The binding of PPARγ to the PPRE regions of the PIK3R1 promoter was analyzed in preadipocytes and mature adipocytes by ChIP assays. After immunoprecipitation, PPRE regions were amplified by PCR. Pre-immune IgG was used as a negative control. M, DNA size markers
The transcriptional activation of PIK3R1 by PPARγ
Our next goal was to identify which of these PPARγ associated sites play a role to regulate the transcriptional activity of PIK3R1. PPRE deletion mutants were constructed for luciferase reporter assays (Fig. 4a). Both PPARγ and RXRα plasmids (P/R) were co-transfected into Cos7 cells to determine the PPARγ-dependency of the PIK3R1 promoter activity. As shown in Fig. 4b, the promoter activity was very low in cells that were transfected with empty reporter vector (pGL3-Basic). In the absence of PPARγ/RXRα heterodimer (pSG), the luciferase activity in both the pGL3-PIK3R1 0.8 kb and pGL3-PIK3R1 1.4 kb vectors was low, but cells expressing exogenous PPARγ/RXRα protein showed increased reporter activity (Fig. 4b). In particular, we observed enhanced luciferase activity in cells that were transfected with the pGL3-PIK3R1 0.8 kb construct containing the PPRE2 domain, suggesting that PPRE2 is necessary domain for PPARγ binding. Next, we investigated the functional activity of PPRE1. A PPRE2 mutant in pGL3-PIK3R1 1.4 kb vector was constructed and measured luciferase activity. In Fig. 4c, the luciferase activity of PPRE2 mutant cells has no difference compared to that of PPRE2 wild type cells, indicating that PPRE1 is also required for the transcriptional activation of PIK3R1 promoter by PPARγ. Taken together, these results suggest that PPARγ regulates the transcriptional activation of PIK3R1 by binding to the PPRE1/2 region.
Identification of the functional PPRE regions in the PIK3R1 promoter. a Schematic structure of various PPRE deletion mutants in the PIK3R1 promoter. The PPRE deletion mutants for the PIK3R1 promoter were designed as pGL3-PIK3R1 0.8 kb and pGL3-PIK3R1 1.4 kb. The pGL3-basic vector was used as a control. b Truncated mutants were co-transfected with empty vector (pSG) or both pSG5-PPARγ and pCMV-RXRα (P/R) into Cos7 cells. At 48 h post-transfection, the luciferase reporter activity was measured. These experiments were conducted as independent experiments in triplicate. Data represent the mean ± SE. **P < 0.01; ***P < 0.005 versus pSG empty vector. c A PPRE2 mutant construct in pGL3-PIK3R1 1.4 kb vector was transfected into Cos7 cells and measured luciferase activity. The data are a representative example of three independent experiments. Data represent the mean ± SE. P/R, PPARγ/RXRα. **P < 0.01 versus pSG empty vector
Discussion
PPARγ and PI3K/AKT are important regulators of adipogenesis and insulin signaling. PPARγ activation is regulated by the protein kinase A (PKA) and AKT signaling pathways [7, 13]. The overexpression of PPARγ does not stimulate adipogenesis in cells silencing AKT protein. The cAMP-induced transcriptional activation of PPARγ disappears in cells exposed to PI3K inhibitor or AKT1 knockdown. These studies indicate that the PPARγ signaling pathway is PI3K/AKT protein-dependent in adipocytes. The activation of PPARγ regulates the expression of insulin signaling cascade-related genes. The phosphorylation of insulin receptor substrate 1 (IRS1), IRS2, and AKT proteins in adipocytes is induced by the PPARγ agonist thiazolidinediones [14, 15]. Insulin is a stimulator of adipocyte differentiation, which regulates glucose uptake and lipogenesis. PPARγ and its ligand induce the uptake of free fatty acids and glucose into adipocytes. In our study, we showed that PPARγ in the presence of insulin synergistically induced the activation of AKT protein and increased PIK3R1 protein expression. In 3T3-L1 cells, the expression of the insulin-responsive glucose transporter GLUT4 is regulated by PPARγ [16]. Once insulin signaling is activated by the PI3K/AKT pathway during adipogenesis, the movement of GLUT4 protein in cells exposed to both insulin and PPARγ is accelerated to the membrane for glucose uptake. In addition to adipocytes, PIK3R1 in muscle, another insulin-responding tissue, is regulated by the PPARα agonist WY-14643 but not by the PPARγ agonist rosiglitazone and 15d-PGJ2 [17]. Although PI3K/AKT signaling is important for insulin action, the regulatory mechanism of PIK3R1 is unique and tissue specific. We found in our study that PPARγ directly regulates PIK3R1 expression in adipocytes, leading to the insulin-induced AKT activation. The increased expression of PPARγ during adipogenesis stimulates the insulin signaling pathway via PIK3R1 activation. Therefore, PPARγ is involved in the insulin-mediated adipogenic pathway in adipocytes. These findings may have important implications concerning the molecular mechanisms of PPARγ-mediated adipogenesis in the insulin-PI3K signaling network.
References
Kershaw EE, Flier JS (2004) Adipose tissue as an endocrine organ. J Clin Endocrinol Metab 89:2548–2556
Galic S, Oakhill JS, Steinberg GR (2010) Adipose tissue as an endocrine organ. Mol Cell Endocrinol 316:129–139
Kawai M, Rosen CJ (2010) PPARγ: a circadian transcription factor in adipogenesis and osteogenesis. Nat Rev Endocrinol 6:629–636
Leto D, Saltiel AR (2012) Regulation of glucose transport by insulin: traffic control of GLUT4. Nat Rev Mol Cell Biol 13:383–396
Duncan RE, Ahmadian M, Jaworski K et al (2007) Regulation of lipolysis in adipocytes. Annu Rev Nutr 27:79–101
Yu W, Chen Z, Zhang J et al (2008) Critical role of phosphoinositide 3-kinase cascade in adipogenesis of human mesenchymal stem cells. Mol Cell Biochem 310:11–18
Xu J, Liao KJ (2004) Protein kinase B/AKT 1 plays a pivotal role in insulin-like growth factor-1 receptor signaling induced 3T3-L1 adipocyte differentiation. Biol Chem 279:35914–35922
Christoffersen CT, Tornqvist H, Vlahos CJ et al (1998) Insulin and insulin-like growth factor-I receptor mediated differentiation of 3T3-F442A cells into adipocytes: effect of PI 3-kinase inhibition. Biochem Biophys Res Commun 246:426–430
Magun R, Burgering BM, Coffer PJ et al (1996) Expression of a constitutively activated form of protein kinase B (c-Akt) in 3T3-L1 preadipose cells causes spontaneous differentiation. Endocrinology 137:3590–3593
Aubin D, Gagnon A, Sorisky A (2005) Phosphoinositide 3-kinase is required for human adipocyte differentiation in culture. Int J Obes (London) 29:1006–1009
Rieusser J, Chambrier C, Bouzakri K et al (2001) The expression of the p85alpha subunit of phosphatidylinositol 3-kinase is induced by activation of the peroxisome proliferator-activated receptor gamma in human adipocytes. Diabetologia 44:544–554
Quandt K, Frech K, Kara H, Wingender E, Werner T (1995) Matlnd and MatInspector: new fast and sensitive tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res 23:4878–4884
Kim SP, Ha JM, Yun SJ et al (2010) Transcriptional activation of peroxisome proliferator-activated receptor-gamma requires activation of both protein kinase A and Akt during adipocyte differentiation. Biochem Biophys Res Commun 399:55–59
Jiang G, Dallas-Yang Q, Li Z et al (2002) Potentiation of insulin signaling in tissues of Zucker obese rats after acute and longterm treatment with PPARγ agonists. Diabetes 51:2412–2419
Smith U, Gogg S, Johansson A et al (2001) Thiazolidinediones (PPARγ agonists) but not PPARα agonists increase IRS-2 gene expression in 3T3-L1 and human adipocytes. FASEB J 15:215–220
Liao W, Nguyen MT, Yoshizaki T et al (2007) Suppression of PPAR-gamma attenuates insulin-stimulated glucose uptake by affecting both GLUT1 and GLUT4 in 3T3-L1 adipocytes. Am J Physiol Endocrinol Metab 293:219–227
Rieusset J, Roques M, Bouzakri K et al (2001) Regulation of p85alpha phosphatidylinositol-3-kinase expression by peroxisome proliferator-activated receptors (PPARs) in human muscle cells. FEBS Lett 502:98–102
Acknowledgments
This research was supported by grants from Basic Science Research Program through the National Research Foundation of Korea (NRF) (No. 2012R1A1B3001134), and by the Next-Generation BioGreen 21 Program (No. PJ008116), Rural Development Administration (RDA), Korea.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kim, YJ., Kim, HJ., Chung, K.Y. et al. Transcriptional activation of PIK3R1 by PPARγ in adipocytes. Mol Biol Rep 41, 5267–5272 (2014). https://doi.org/10.1007/s11033-014-3398-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3398-9