Abstract
The estrogen receptor 1 (ESR1) and Chromosome 6 Open Reading Frame 97 (C6orf97) gene polymorphisms were earlier reported to be associated with osteoporosis in the European cohort. The aim of this study was to investigate the association of four single nucleotide polymorphisms (SNP) with bone mineral density (BMD), fracture, vertebral fracture, bone turnover or 25-hydroxyvitamin D [25(OH)D] in 1,753 randomly selected postmenopausal women in China. Vertebral fracture, BMD of lumbar spine (2–4), femoral neck and total hip were measured respectively. Serum N-terminal procollagen of type 1 collagen (P1NP), β-isomerized type I collagen C-telopeptide breakdown products (β-CTX) and 25(OH)D3 were also determined. Binary logistic regression revealed significant associations between fracture risk with rs1999805 (P = 0.041, OR 1.633, 95 %CI 1.020–2.616) and rs6929137 (P = 0.005, OR 1.932, 95 %CI 1.226–3.045) in recessive model. Significant association was also observed between vertebral fracture risk and rs1038304 (P = 0.039, OR 0.549, 95 %CI 0.311–0.969) in recessive model. Liner regression analyses showed that only the CC group of rs4870044 was significantly associated with total hip in dominant model (P = 0.034). Our findings suggest that ESR1 and C6orf97 gene polymorphism is associated with fracture and vertebral fracture risk in Chinese postmenopausal women.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Osteoporosis is a systemic bone disease which is characterized by low bone mass, damaged bone micro-structural, leading to increased bone fragility and trend to fracture [1]. It is a multifactorial complex disease which is affected by genetic and environmental factors. Bone markers and bone mineral density (BMD) were important indicators for the risk of fracture, especially vertebral fracture [2, 3].
Genome-wide association study (GWAS) has found several new single-nucleotide polymorphism (SNP) sites that were associated with BMD. Recently, in order to find sensitive genetic markers of osteoporosis, generous researches have been carried out on candidate genes involved in osteoporosis for drug prevention and treatment of osteoporosis. The related genes include vitamin D receptor (VDR) gene on chromosome 12q13-14 [4, 5], estrogen receptor (ESR1) gene on chromosome 6q25.1 [6], type 1 collagen alpha 1 (COL1A1) gene on chromosome 17q21.3-q22 [7, 8], and LDL receptor-like protein-5 (LRP5) gene on chromosome 11q13.4 [9, 10].
Meta-analysis on osteoporosis showed that polymorphisms of ESRl, COLlAl, VDR genes could only explain a small part of the genetic basis of osteoporosis. GWAS has been firstly carried out in SNPs to explore the most common osteoporosis-related phenotypes in 1,141 participants from Framingham study [11]. Ten of the common phenotypes were reported to be correlated to osteoporosis. Then GWAS was then used in Icelandic population by Unnur Styrkarsdottir [12] and in Western Europe by Richards JB [13]. These GWAs found some SNPs within or close to the SNPs which were previously thought to be associated with BMD, such as loci located in ESRl and LRP5 genes.
ESR1 gene plays an important role in bone growth and maintenance of bone mass by regulating metabolism and acquisition of peak mass of bone as well as inhibiting bone loss. A large number of studies have shown that estrogen is essential for bone homeostasis maintaining. Three genetic markers, PvuII (T>C), XbaI (A>G) and TA repeats of the promoter region, were located in intron 1. ESR1 gene was reported to be associated with BMD of white women in Scotland [6]. The haplotype “PvuII-XbaI” could reduce BMD of the hip and speed up the bone loss of the femur and collarbone.
A plurality of SNP loci on 6q25.1 region of chromosome was associated with BMD. Previous report has shown that ESR1 gene polymorphism was associated with osteoporosis and BMD [14–16]. A haplotype of ESR1 was reported to be associated with reduced hip BMD and increased rates of femoral neck bone loss in postmenopausal women [6]. This area includes not only ESRl gene but also upstream C6orf97 gene. And multiple loci of the C6orf97 gene were found be strongly associated with BMD. Some of the loci have been verified to be associated with BMD in cohorts of Denmark, Australia, Europe and US [13, 17].
Nevertheless, the association of these loci with BMD and fracture in Chinese cohort has not been reported. In this study, association between BMD and fracture and four loci (rs1999805, rs4870044, rs1038304, and rs6929137) in 6q25.1 region were investigated in postmenopausal women in China. The four loci were previous found to be strongly associated with BMD in Iceland cohort. Our aim was to analyze the distribution of these loci in Chinese cohort to explore the relationship of which with BMD and vertebral fracture.
Materials and methods
Patients
Postmenopausal female participants were from a Peking Vertebral Fracture (PK-VF) cohort that was selected randomly from seven districts in Beijing, China [18]. Samples were stratified into four age groups according to the community population (50–59, 60–69, 70–79, and ≥80 years old). Finally, 1,753 participants, without diseases or drug use that affecting bone metabolism, were enrolled in this study. Population sampling and data collection were performed by the same standard. The exclusion criteria were as follows: (1) severe liver and kidney disease; (2) rheumatic disease or rheumatoid arthritis, connective tissue disease; (3) metabolic or genetic bone diseases, including hypothyroidism or hyperthyroidism, Paget disease, osteogenesis imperfecta, osteomalacia, etc.; (4) gastrointestinal disease, including chronic diarrhea, peptic ulcer, Crohn’s disease, etc.; (5) Application of steroid hormone or anticonvulsants for 6 months or more; (6) previous long-termuse (6 months or more) or currently using drugs for osteoporosis treatment, including bisphosphonates, estrogen, calcitonin, and selective estrogen receptor modulators. The study was approved by the Ethic Committee of Peking Union Medical College Hospital (PUMCH). Informed consent forms were obtained from all participants before entering the study. Clinical data were obtained from VF-risk factors questionnaire which was performed by uniformly trained researchers.
Bone marker determination
Fasting blood samples were taken and the serum was isolated by centrifuge at 4,000 rpm for 5 min. Serum bone markers including 25-hydroxy-Vitamin D3 (25-OH-D3), cross linked C-telopeptide of type 1 collagen (CTX) and N-terminal procollagen of type I collagen (P1NP) were determined by Roche electrochemiluminescence system (Roche Diagnostics, Switzerland).
Bone mineral density (BMD) examination
BMD of total hip, femoral neck (FN) and lumbar spine (LS) were measured with dual energy X-ray absorptiometry (DXA) machines of either Lunar DPX (General Electric Healthcare, USA) or Norland (USA). Calibration with manufacturer’s phantom was performed every day before the examination. Calibration equations between the two machines were as follows [19]:
According to the WHO definition [20], T-Score ≤ −2.5 standard deviation (SD) is considered as osteoporosis, and −1.0SD > T-Score > −2.5SD is considered as osteopenia. Accordingly, participants with T-Score ≥ −1.0SD were considered normal bone mass.
Evaluation of vertebral fractures
The participants received lumbar spine X-ray examination (T4 to L5) which was performed by uniformly trained technicians. The results were evaluated according to the Genant semi-quantitative method [21]. Normal shape and size of the vertebral of T4 to L5 under standard lateral X-ray was defined as grade 0. Grade 1 to grade 3 was characterized by reduced height and projected area of the vertebral (20–25 and 10–20 % for grade 1, 26–40 and 21–40 % for grade 2, and both more than 40 % for grade 3, respectively). Vertebral fracture was defined as visible vertebral fractures on X-ray film.
Genomic DNA extraction and quantification
Genomic DNA was extracted using a blood genomic DNA extraction kit (TIANGEN CO. LTD, Beijing, China) according to the manufacture’s instruction. Purity of the obtained DNA was evaluated by OD260/OD280 ratio using a spectrophotometer (UV-1700, SHIMADZU, Japan). An OD260/OD280 ratio between 1.6 and 2.0 was considered high purity of DNA. Quantification of genomic DNA was performed by the following formula: DNA concentration (μg/ml) = OD260 × 50 × dilution factor. Distilled water was used as the blank control. Based on the concentration, DNA samples were standardized to 5 ng/μl and stored at −20 °C.
SNP determination
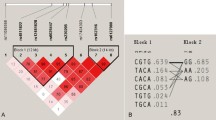
SNP data of the 6q25.1 region was obtained from Hapmap website (http://www.hapmap.org). Linkage disequilibrium region of genes were defined by the SNP loci by Haploview 4.1 software. Figure 1 shows the genetic linkage disequilibrium region of C6orf97 gene, in which darker color means higher degree of linkage. Tag SNP was selected by the Tagger options which represent the maximum amount of genetic information in the chromosomal region. Finally, four SNP loci were detected: rs6929137, rsl999805, rs4870044, and rs1038304. Details of the four loci were shown in Table 1.
Linkage disequilibrium in C6orf97 gene located in 6q25.1 region of chromosome. Data of C6orf97 gene located in 6q25.1 region was downloaded from http://www.hapmap.org (on November 21, 2013) and the linkage disequilibrium was determined by Haploview 4.1 software. Darker intersecting means higher the degree of linkage disequilibrium
SNP detection was performed using TaqMan allelic discrimination assay (Applied Biosystems, USA).The sequences of each probe were synthesized by Applied Biosystems, withVlC/FAM fluorophore at the 3′end and non-fluorescent quencher (NFQ) at the 5′ end. The details were as follows: rs1999805: CGTACCCAGAAAGCTCTTAGATTGT[A/G]AATGCTGAGGGAACACTGGGAGATT; rs4870044: GCTATGTCATTTGTGGTATAGGAGA[C/T]GTTGAAGGGTCTTCAGGTAGAAATA; rs1038304: GGTTTCTAGTTTATGAGTTCAGCAG[A/G]TATTTGACTCTGGATTCCTCAATTT; rs6929137: GAAAGCTGAGAAAAAGCTCATGTCT[A/G]TCAAGTCAGAACTGGATACCACAGA.
The PCR mixture (5 μl final volume per reaction) was prepared according to the manufacturer’s protocol. Reactions were performed in a 384-well plate on a Real-Time PCR system of ABI Prism 7900 (Applied Biosystems, USA). Amplifications were performed with 2 min at 50 °C, 10 min at 95 °C, followed with 45 cycles of 15 s at 92 °C, 60 s at 60 °C.
Hardy–Weinberg equilibrium test
In this study, osteoporosis and osteopenia is defined as the group of low bone mass, and normal bone mass is defined as the control group. Participants with or without vertebra fracture were defined as the case group and the control group, respectively. Bone markers above or below the 50th percentile were defined as the case group and the control group, respectively. Genotype frequencies analysis of the case group and the control group were performed in order to investigate Hardy–Weinberg genetic equilibrium in each SNP.
Statistical analysis
Genetic analysis including Hardy–Weinberg equilibrium test, minor allele frequencies and haplotype frequencies were performed using Haploview software (Cambridge, MA). According to the number of minor allele or copies of haplotype, genotypes and haplotypes were divided into three models, including additive model (code 0, 1 and 2), dominant model (code 0, 1 and 1), and recessive model (code 0, 0 and 1). Statistical analysis was performed with SPSS 15.0 software (SPSS® Statistics, Chicago), using the method of t test, analysis of variance (ANOVA), analysis of covariance, and Chi square test. All measurement data are normally distributed, homogeneity of variance was tested before ANOVA. Data from the experiments are expressed as mean ± standard deviation. After adjusted for age, BMI and YSM, associations between fractures (fracture and vertebral fracture) with genotype or haplotype were analyzed using binary logistic regression. After the same adjustment, associations between genotypes with BMD T-scores of LS, FN and total hip were analyzed using linear regression. Differences on β-CTX, P1NP (adjusted for age and YSM), and 25(OH)D (adjusted for age) in different genotype groups were calculated with general linear model ANOVA (GLM-ANOVA). A P value of <0.05 was considered significant.
Results
Hardy–Weinberg equilibrium test on SNP loci of 6q25.1 region
Table 2 shows the result of Hardy–Weinberg equilibrium test on the total samples, indicating that the SNP loci of 6q25.1 region were in line with Hardy–Weinberg law (P > 0.05). Besides, the same method was used between the group of low bone mass and normal group, with or without vertebral fracture, high or low bone turnover. All the test showed P values above 0.05, which indicated that all the SNP loci in our study were in line with Hardy–Weinberg law. This means that the subjects chosen in our study could represent the whole population. The MAF values of rs1999805, rs4870044, rs1038304 and rs6929137 were 0.32 (A), 0.21 (C), 0.48 (G) and 0.32 (A).
Association analysis of genotypes of the SNP loci in 6q25.1 region
To investigate the relation of SNP loci in 6q25.1 to fracture and vertebral fracture risk, binary logistic regression of genotypes in relation to fracture and vertebral fracture risk were performed in participants after age matching. As shown in Table 3, significant associations were shown between fracture risk with rs1999805 (P = 0.041, OR 1.633, 95 %CI 1.020–2.616) and rs6929137 (P = 0.005, OR 1.932, 95 %CI 1.226–3.045) in recessive model after adjusted for BMI and YSM. In addition, significant association was shown between vertebral fracture risk with rs1038304 (P = 0.039, OR 0.549, 95 %CI 0.311–0.969). However, other SNPs did not show any significant association with fracture or vertebral fractures (P > 0.05).
Linear regression was performed to further investigate the relation of SNP loci in 6q25.1 to LS, FN and total hip. The results showed that only the CC group of rs4870044 was significantly related with total hip in dominant model (P = 0.034). Other groups of the four SNPs did not show any significant relation with LS, FN or total hip (P > 0.05) (Table 4).
Results of GLM–ANOVA showed that the level of P1NP of participants with AA genotype of rs1038304 was significantly higher compared with other genotypes of the SNP loci (P = 0.028), while none of the other genotype of SNP loci in 6q25.1 region showed significant relation with β-CTX, P1NP or 25(OH)D3 (Table 5).
Haplotype analysis
The haplotype block was defined by Haploview4.1 software and the constructed haplotypes, rs1038304 and rs6929137 in the same linkage disequilibrium region (LD block1), were shown in Fig. 2 (D′ = 0.954, r2 = 0.467). The haplotypes were AG, GA and GG, with AG haplotype the most common (51.4 %), followed with GA haplotype (31.3 %). All the three haplotypes showed no difference between case and control groups of vertebral fracture, the same negative results were shown in case and control groups of low BMD (P > 0.05, data not shown).
Discussion
This study chose four new SNP loci which were found to be related with BMD by GWAS. We investigated the possible association of SNP and osteoporosis phenotypes in Chinese postmenopausal women. This study found that three genotypes of rs1038304 were related with P1NP. Out of the three genotypes, AA genotype showed significant difference with the other two by pairwise comparisons analyze. The difference remained significant after adjustment for age, height, body weight, and YSM. Since P1NP is a specific marker of bone formation, a prudent inference was made that rsl038304 may be associated with bone formation. Some similar results were shown in other previous studies. Heemstra et al. recently found that Thr92Ala gene polymorphism of deiodinase type 2 was related to P1NP level [22]. Another study have explored the relation of P1NP or CTX with some SNP loci within RANKL/RANK/OPG pathway in European male [23], A locus of Interleukin-6 (IL-6) was also found to be related with serum CTX level in postmenopausal women [24]. But so far rare evidence has suggested the heritability of bone markers.
We analyzed the relation of BMD and allele frequencies as well as genotypes. None of the four SNPs showed significant association with BMD or vertebral fracture. In order to enlarge the difference between genotypes, dominant-recessive model was established. Significant association was shown between three of the SNPs with facture or vertebral fracture in recessive model (P < 0.05). These results indicated that some of the loci which were associated with BMD or vertebral fractures in Eurpean cohort may be associated with BMD or vertebral fractures in Chinese cohort. A study performed in 1,000 white Americans by Zhang et al. has validated some positive loci which were found by GWAS. They found that rs4870044 was associated with femur neck and vertebral BMD in women (P = 1.8 × 10−4 and 1.3 × 10−2, respectively) [13]. These findings suggested the potential role of race in the difference of association between gene loci and BMD or vertebral fracture.
Further analysis showed that rs6929137 and rs1038304 were included in a LD block with the length of about 3 kb (r2 = 0.467). AG, GA and GG haplotypes were constructed, and the most common haplotype was AG (51.4 %). Analysis by liner regression showed that only the CC group of rs4870044 was significantly related with total hip in dominant model (P = 0.034). Other groups of the four SNPs did not show any significant relation with LS, FN or total hip (P > 0.05). Recently, another study has explored the association between rs4870044 and BMD loss [25]. Results of GLM-ANOVA showed that the level of P1NP of participants with AA genotype of rs1038304 was significantly higher compared with other genotypes of the SNP loci (P = 0.028). This indicated that the AA genotype of rs1038304 may be associated with bone formation. The potential role of these SNPs needs further study due to the limit reports.
Ethnic differences may play a role in the relationship between the four SNPs and BMD or vertebral fracture. The Hapmap indicated the significant differences of gene distribution between Caucasians and Beijing Han population in the same SNP loci. For instance, MAF of rs1999805 in Caucasian, Tokyo, and Beijing was 0.42 (G), 0.34 (A), and 0.27 (A), respectively. In this study, we found the MAF of this locus was 0.32 (A). Differences in the distribution of genes resulted in different results of gene polymorphism in different races, which may explain why the four SNP loci in this study were highly correlated with BMD in Europe and US but showed remarkable difference in Chinese cohort.
Osteoporosis is a multifactorial disease, and genetic factor was only a part of the reason for occurrence and development of osteoporosis. Furthermore, postmenopausal osteoporosis has recently been reported to be associated with a plurality of gene polymorphisms within multiple chromosomal regions, such as GALNT3 [26], ESR1 [27], VDR and COL1A1 [28, 29]. A considerable number of confounding factors need to be considered to explore whether SNP or single gene could eventually affect the phenotype. Execution of gene function requires transcription, translation, and a series of processes, including the interaction between genes and proteins. Our study indicated that some of these loci were indeed associated with osteoporosis in the Chinese population but the correlation is not significant. The sample size in this study may be an influencing factor of the results.
In summary, genotype of rs1038304 within 6q25.1 region of the chromosome was associated with high P1NP level in China Beijing postmenopausal women. The CC genotype of rs4870044 was significantly related with total hip in dominant model. The AA genotype of rs1038304 may be associated with bone formation. A limitation of our study comes from its small patient population; hence larger studies are required to validate our findings.
Accordingly, we suggest that in addition to the clinical risk factors, the variants in ESR1 and C6orf97 are susceptibility genetic loci for osteoporotic fracture in postmenopausal women in China. It is imperative that future studies have to intuitively query the underlying mechanisms responsible for association between osteoporotic fractures and protective loci/genetic risk. Such endeavors have to GWASs, large-scale association studies, as well as meta-analyses. Pending further validation, clinical decision making and risk prediction in postmenopausal women might include ESR1 and C6orf97 genotyping in China, and elsewhere across the globe.
References
Genant HK, Cooper C, Poor G et al (1999) Interim report and recommendations of the world health organization task-force for osteoporosis. Osteoporos Int 10:259–264
Melton LJ 3rd, Khosla S, Atkinson EJ et al (1997) Relationship of bone turnover to bone density and fractures. J Bone Miner Res 12:1083–1091
Gerdhem P, Ivaska KK, Alatalo SL et al (2004) Biochemical markers of bone metabolism and prediction of fracture in elderly women. J Bone Miner Res 19:386–393
Grundberg E, Lau EM, Pastinen T et al (2007) Vitamin D receptor 3′ haplotypes are unequally expressed in primary human bone cells and associated with increased fracture risk: the MrOS Study in Sweden and Hong Kong. J Bone Miner Res 22:832–840
Abrams SA, Griffin IJ, Hawthorne KM et al (2005) Vitamin D receptor Fok1 polymorphisms affect calcium absorption, kinetics, and bone mineralization rates during puberty. J Bone Miner Res 20:945–953
Albagha OM, Pettersson U, Stewart A et al (2005) Association of oestrogen receptor alpha gene polymorphisms with postmenopausal bone loss, bone mass, and quantitative ultrasound properties of bone. J Med Genet 42:240–246
Niu T, Chen C, Cordell H et al (1999) A genome-wide scan for loci linked to forearm bone mineral density. Hum Genet 104:226–233
Ralston SH, Uitterlinden AG, Brandi ML et al (2006) Large-scale evidence for the effect of the COLIA1 Sp1 polymorphism on osteoporosis outcomes: the GENOMOS study. PLoS Med 3:e90
van Meurs JB, Trikalinos TA, Ralston SH et al (2008) Large-scale analysis of association between LRP5 and LRP6 variants and osteoporosis. JAMA 299:1277–1290
Lau HH, Ng MY, Ho AY et al (2005) Genetic and environmental determinants of bone mineral density in Chinese women. Bone 36:700–709
Kiel DP, Demissie S, Dupuis J et al (2007) Genome-wide association with bone mass and geometry in the Framingham Heart Study. BMC Med Genet 8(Suppl 1):S14
Styrkarsdottir U, Halldorsson BV, Gretarsdottir S et al (2008) Multiple genetic loci for bone mineral density and fractures. N Engl J Med 358:2355–2365
Zhang YP, Deng FY, Chen Y et al (2010) Replication study of candidate genes/loci associated with osteoporosis based on genome-wide screening. Osteoporos Int 21:785–795
Sano M, Inoue S, Hosoi T et al (1995) Association of estrogen receptor dinucleotide repeat polymorphism with osteoporosis. Biochem Biophys Res Commun 217:378–383
Kobayashi S, Inoue S, Hosoi T, Ouchi Y, Shiraki M, Orimo H (1996) Association of bone mineral density with polymorphism of the estrogen receptor gene. J Bone Miner Res 11:306–311
van Meurs JB, Schuit SC, Weel AE et al (2003) Association of 5′ estrogen receptor alpha gene polymorphisms with bone mineral density, vertebral bone area and fracture risk. Hum Mol Genet 12:1745–1754
Styrkarsdottir U, Halldorsson BV, Gretarsdottir S et al (2009) New sequence variants associated with bone mineral density. Nat Genet 41:15–17
Zhao J, Xia W, Nie M et al (2011) The levels of bone turnover markers in Chinese postmenopausal women: peking vertebral fracture study. Menopause 18:1237–1243
Kocks J, Ward K, Mughal Z, Moncayo R, Adams J, Hogler W (2010) Z-score comparability of bone mineral density reference databases for children. J Clin Endocrinol Metab 95:4652–4659
Assessment of fracture risk and its application to screening for postmenopausal osteoporosis. Report of a WHO Study Group. World Health Organ Tech Rep Ser 1994, 843:1–129
Genant HK, Wu CY, van Kuijk C, Nevitt MC (1993) Vertebral fracture assessment using a semiquantitative technique. J Bone Miner Res 8:1137–1148
Heemstra KA, Soeters MR, Fliers E et al (2009) Type 2 iodothyronine deiodinase in skeletal muscle: effects of hypothyroidism and fasting. J Clin Endocrinol Metab 94:2144–2150
Roshandel D, Holliday KL, Pye SR et al (2010) Genetic variation in the RANKL/RANK/OPG signaling pathway is associated with bone turnover and bone mineral density in men. J Bone Miner Res 25:1830–1838
Ferrari SL, Garnero P, Emond S, Montgomery H, Humphries SE, Greenspan SL (2001) A functional polymorphic variant in the interleukin-6 gene promoter associated with low bone resorption in postmenopausal women. Arthritis Rheum 44:196–201
Yao S, Sucheston LE, Smiley SL et al (2011) Common genetic variants are associated with accelerated bone mineral density loss after hematopoietic cell transplantation. PLoS ONE 6:e25940
Li N, Wang X, Jiang Y et al (2013) Association of GALNT3 gene polymorphisms with bone mineral density in Chinese postmenopausal women: the peking vertebral fracture study. Menopause 18:1237–1243
Cheng BH, Wang TH, Kang HY et al (2013) Association between single nucleotide polymorphisms of the estrogen receptor 1 and receptor activator of nuclear factor kappa B ligand genes and bone mineral density in postmenopausal Taiwanese. Taiwan J Obstet Gynecol 52:197–203
Marozik P, Mosse I, Alekna V et al (2013) Association between polymorphisms of VDR,COL1A1, and LCT genes and bone mineral density in belarusian women with severe postmenopausal osteoporosis. Medicina (Kaunas) 49:177–184
Gonzalez-Mercado A, Sanchez-Lopez JY, Regla-Nava JA et al (2013) Association analysis of vitamin D receptor gene polymorphisms and bone mineral density in postmenopausal Mexican-Mestizo women. Genet Mol Res 12:2755–2763
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Luo, L., Xia, W., Nie, M. et al. Association of ESR1 and C6orf97 gene polymorphism with osteoporosis in postmenopausal women. Mol Biol Rep 41, 3235–3243 (2014). https://doi.org/10.1007/s11033-014-3186-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-014-3186-6