Abstract
Changes in the expression profiles of microRNAs (miRNAs) have been found in many cancers. The study was aimed to investigate the expression of miR-25, miR-223, and miR-375 in the serum of patients with esophageal squamous cell carcinoma (ESCC) and its effect on survival outcome. We examined the expression levels of miR-25, miR-223, and miR-375 in 20 pairs of ESCC cancer and matched paracancerous tissues, serum samples from 94 healthy volunteers and 194 patients with ESCC using quantitative reverse transcription polymerase chain reaction, and analyzed the relationship between expressions of serum miR-25, miR-223, and miR-375 and ESCC clinicopathological parameters as well as survival. Expressions of miR-25 and miR-223 were significantly increased in ESCC tissues compared with paracancerous tissues (P = 0.008 and 0.009, respectively), whereas the expression of miR-375 was significantly decreased in ESCC tissues compared with paracancerous tissues (P = 0.006). Expressions of serum miR-25 and miR-223 were significantly higher in ESCC patients than those in healthy controls, and, inversely, expression of serum miR-375 was significantly lower in ESCC patients than those in healthy controls (P = 0.007). High expression of serum miR-25 was significantly associated with lymph node metastasis (P = 0.01). Survival analysis showed that high expression of serum miR-223 and low expression of serum miR-375 were associated with poor survival in ESCC patients [hazard ratio (HR) = 1.717, 95 % confidence intervals (CI) 1.139–2.588, P = 0.01; HR = 1.750, 95 % CI 1.111–2.756, P = 0.016, respectively). Furthermore, Patients with high miR-223 and low miR-375 expression had higher risk of death than those with low miR-223 and high miR-375 expression (HR = 3.599, 95 % CI 1.800–7.195, P = 2.92 × 10−4). In conclusion, miR-25, miR-223, and miR-375 were abnormally expressed in ESCC tissues and sera. Serum miR-223 and miR-375 are potential prognostic biomarkers for ESCC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Esophageal carcinoma is one of the most common malignancies worldwide. Approximately 482,300 newly diagnosed cases of esophageal carcinoma were reported worldwide in 2008, accounting for ~3.8 % of all new cancer cases [1]. In china, esophageal squamous cell carcinoma (ESCC) is the predominant histological subtype, accounting for 90 % of the total esophageal carcinoma [1]. Major therapies for esophageal carcinoma now include surgery, chemotherapy, and radiotherapy. Although survival and functional recovery rates have remarkably improved as a result of the continuous advancement of diagnostic technologies and therapies, the outcome is far from satisfactory with 10–30 % overall 5-year survival rate [2, 3]. Numerous factors influence the prognosis of esophageal carcinoma, such as clinicopathological features [4–6] and abnormal expression of genes and/or proteins [7–10]. However, molecular markers that are of clinical application value are quite limited [10]. Further studies are needed to demonstrate or search for markers that can serve as reliable evidence to determine the efficacy and outcome of esophageal carcinoma treatment.
MicroRNAs (miRNAs) are a class of small non-coding RNA molecules that can control gene expression post-transcriptionally. These molecules are important in cell growth, proliferation, metabolism, and apoptosis. In recent years, the relationship between miRNAs and human tumors has attracted much attention. The imbalanced expression of several key miRNAs is an important factor in the occurrence and development of cancer [11–13]. Therefore, numerous studies have been using gene chip technology to search for miRNAs with specific expression in cancer tissues by comparing cancer and paracancerous tissues. miRNAs are presumed to regulate nearly one-third of human gene expression [14, 15]. Abnormal patterns of miRNA expression have been observed in lung [16], colorectal [17, 18], esophageal [19, 20], and kidney and bladder [21] cancers. Furthermore, aberrant miRNA expression is closely related to metastasis and prognosis of cancer [22–25]. It has been shown that circulating miRNAs can be steadily detected to suggest the cancer status [25–27]. Thus, circulating miRNAs can be considered as a valuable marker for cancer monitoring. Considering the ease of blood sampling, circulating miRNAs hold better potential for future application as a new cancer biomarker.
Recent studies have reported diverse abnormal expression of miR-25 [28, 29], miR-223 [30–32], and miR-375 [17, 19, 33–36] in various types of cancer, including ESCC [29, 32, 36]. MiR-25 and miR-223 function as an oncogene or tumor suppressor in different cancer types [18, 29, 30, 33, 37–39]. MiR-25 represses migration and invasion of ESCC cells by targeting E-cadherin [29]. Overexpression of miR-223 inhibits FBXW7 expression in ESCC cells and then leads to abnormal accumulation of c-Myc and c-Jun proteins [32]. miR-375 acts as a tumor suppressor by targeting several oncogenes in cancer cells [40]. However, numerous previous studies only compared cancer and normal tissues. Since tissue sampling is difficult, especially for patients in advanced stages, which has limited the wide application of miRNA. Furthermore, little data have been reported to date regarding the relationship between circulation miRNAs and prognosis of esophageal cancer patients. Therefore, in the present study, we used quantitative reverse transcription polymerase chain reaction (qRT-PCR) to investigate the expressions of miR-25, miR-223, and miR-375 in ESCC patients. We also investigated the relationship between the expression levels of serum miR-25, miR-223, and miR-375 and the clinical factors as well as analyzed their effects on survival in ESCC patients.
Materials and methods
Patients
With informed consents, we enrolled 20 cases of ESCC patients who underwent surgeries in Zhongshan Hospital between April 2006 and March 2012. Surgical specimens were taken post-operationally for each patient within 30 min, using blades to cut off a piece each of esophageal cancer and paracancerous tissues (2 cm from the cancer tissue). Fresh specimens obtained by surgical ablation were immediately placed and stored in liquid nitrogen. We collected 2 mL peripheral blood from each of 194 ESCC patients and 98 healthy controls. All patients were pathologically diagnosed as having ESCC using surgical specimens or biopsies. None of the ESCC patients had received any anticancer treatment prior to sampling. Approval was obtained from the Ethics Committee of Zhongshan Hospital prior to specimen collection. Table 1 shows the clinical profiles of patients.
Quantitative real-time PCR
Total RNA was extracted from tissue and serum samples, according to the manual of miRNAVanaTM PARISTM (Ambion, TX, USA). RNA (1 μL) concentration was quantified using NanoDrop ND-1000 Spectrophotometer (NanoDrop, Wilmington, DE, USA). RNA was transported and stored in a refrigerator at 80 °C.
TaqMan microRNA Reverse Transcription Kit (Applied Biosystems, CA, USA) was used in reverse transcription. The reaction system was 15 μL, containing 100 ng total RNA, 1.5 μL 10× reverse transcription buffer, 0.15 μL 100 mM deoxyribonucleotide triphosphates, 1.0 μL reverse transcriptase, 0.19 μL RNase inhibitor (20 U/μL), and 3.0 μL specific miRNA primer. Reactions were performed at 16 °C for 30 min, 42 °C for 30 min, and finally at 85 °C for 5 min. The 20 μL reaction mixtures for qRT-PCR included 2 μL cDNA, 10 μL 2 × Universal PCR Master Mix, 1.0 μL TaqMan miRNA assay, and 7 μL nuclease-free water. qRT-PCR was performed on ABI7500 fast real-time PCR system (Applied Biosystems, CA, USA) under the following reaction conditions: 50 °C for 2 min and 95 °C for 10 min, followed by 40 cycles: at 95 °C for 15 s and at 60 °C for 1 min. Each reaction was repeated thrice. The expression levels of miRNAs in tissue and serum samples were normalized to U6 small nuclear RNA (RNU6B) and miR-16 [41], respectively, and were calculated using the equation 2−ΔΔCt. The ΔCt was calculated by subtracting the average Ct value of corresponding reference gene from the average Ct value of the miRNAs of interest. The ΔΔCt was then calculated by subtracting the ΔCt of paracanerous tissue or the average expression of healthy volunteers from ΔCt of ESCC patients. The fold change in gene expression was calculated with the equation 2−ΔΔCt. The expression levels of miRNAs were converted into dichotomous variables by splitting the samples into two classes (high and low expression), using the respective mean level expression of miRNA as a cutoff [42].
Statistical analysis
Data analysis was performed with SPSS 17.0 (SPSS Inc, Chicago, USA). P < 0.05 was considered statistically significant. Rank-sum test was used to compare the differences in the expression of serum miR-25, miR-223, and miR-375 between ESCC patients and healthy controls. χ 2-test was used to determine the relationship between the expression of these serum miRNAs and the ESCC clinicopathological parameters. Receiver-operating characteristic (ROC) curves and the area under the ROC curve (AUC) were used to assess the predictive power of individual miRNAs. Kaplan–Meier method and log-rank test were used to investigate the association between the expression of the three serum miRNAs and the survival in ESCC patients. Cox regression model was used to test the independence of risk factors. The end point was overall survival from the time of initial histological diagnosis.
Results
MiR-25, 223, and 375 in primary ESCC tissues
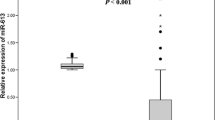
Differential expression of miR-25, miR-223, and miR-375 in cancer and paracancerous tissues were statistically significant (P = 0.008, 0.009 and 0.006, respectively) (Fig. 1). The expression levels of miR-25 and miR-223 were increased, whereas the expression level of miR-375 was decreased in ESCC tissues (fold change = 2.66, 3.42 and 2.71, respectively).
Expression of serum miR-25, 223, and 375 in ESCC patients
We hypothesized that the expression levels of miR-25, miR-223, and miR-375 in primary ESCC tissues would influence serum levels of miR-25, miR-223, and miR-375 in ESCC patients. We first compared the expression levels of miR-25, miR-223, and miR-375 in 20 pairs of ESCC tissues and sera. There were statistically significant correlations between tissue and serum levels of miR-25, miR-223, and miR-375 (correlation coefficients ranged from 0.579 to 0.663, all P < 0.01) (Fig. S1). We further investigated the serum miRNA expression in 194 ESCC patients and 98 healthy controls. We found statistically significant differences between ESCC patients and healthy controls in expression levels of miR-25, miR-223, and miR-375 (P = 0.009, 0.001 and 0.007, respectively) (Fig. 1).
Relationship between expression of serum miR-25, miR-223, and miR-375 and clinicopathological characteristics in ESCC patients
We also analyzed the relationship between serum miR-25, miR-223, and miR-375 expression and clinicopathological features in ESCC patients in order to better understand their potential roles in the development and progression of ESCC. High expression of serum miR-25 was significantly associated with lymph node metastasis (P = 0.01). The AUC based on serum miR-25 was 0.593 (Fig. 2a). No other significant differences were found between the expression levels of serum miR-25, miR-223 and miR-375, and clinicopathological characteristics (all P > 0.05) (Table 2).
ROC analyses of miR-25, miR-223 and miR-375 based on serum expression. a Serum miR-25 yielded AUC of 0.593 (95 % CI 0.511–0.676) with 47.1 % sensitivity and 71.6 % specificity in discriminating ESCC patients with lymph node metastasis. b ROC curve for 5-year survival for serum miR-223. c ROC curve for 5-year survival for serum miR-375. d ROC curve for 5-year survival for combined miR-223 and miR-375
Survival analysis
The median survival for the whole group was 31.3 months. The overall survival rates at 1, 3, and 5 years were 85.8, 42.5, and 29.4 %, respectively. Patients with upregulated miR-223 expression had significantly shorter median survival time (MST) than those with low miR-223 expression (26.3 vs. 36.9 months; P = 0.007, log-rank test; Fig. 3a). By contrast, MST was significantly longer for those with upregulated miR-375 expression than those with low miR-375 expression (39.3 vs. 27.8 months; P = 0.016, log-rank test) (Fig. 3b). The AUC based on serum miR-223 and miR-375 expression was 0.734 and 0.720, respectively (Fig. 2b, c). No significant association was observed between miR-25 expression and survival (P = 0.525). Multifactor Cox analysis showed that high serum miR-223 expression and low serum miR-375 expression were independent predictors of survival in ESCC patients [Hazard ratio (HR) = 1.717, 95 % confidence intervals (CI) 1.139–2.588, P = 0.010; HR = 1.750, 95 % CI 1.111–2.756, P = 0.016, respectively) (Table 3).
To perform a comprehensive analysis of the two markers, we assigned ESCC patients to three groups: group one, patients with low miR-223 and high miR-375 expression; group two, patients with high miR-223 or low miR-375 expression; group three, patients with high miR-223 and low miR-375 expression. Patients in group one had longer MST than those in group two or three (P = 0.002) (Fig. 3c). In the multivariate Cox-regression model, patients in group two had a cancer-related death risk 2.527 times higher (95 % CI 1.268–5.039, P = 0.008), and those in group three had a cancer-related death risk 3.599 times higher (95 % CI 1.800–7.195, P = 2.92 × 10−4) than those in group one. The AUC based on combined miR-223 and miR-375 was 0.745 (Fig. 2d).
Discussion
The pathogenesis of esophageal cancer remains unknown. With the progression of cancer research, no or little gene mutations were found in certain cancers to date. Moreover, miRNAs play vital roles in post-transcriptional regulation in many biological processes. Therefore, researchers are increasingly shifting their focus on the relationship between miRNAs and tumors. In the present study, we used qRT-PCR to determine the expressions of serum miR-25, miR-223, and miR-375 in ESCC patients and healthy controls. We found that serum miR-223, miR-25 and miR-375 expression are potential biomarkers for the prognosis of ESCC patients.
miR-25 and miR-223 are upregulated in many types of cancer [29, 30, 37, 43, 44], promoting cancer cell proliferation, migration and invasion [29, 30, 37, 45]. However, miR-25 may act as tumor suppressor, and inhibits proliferation of colon cancer [18] and anaplastic thyroid carcinoma cells [38]. These results indicates that miRNAs plays distinct roles under different cellular. miR-375 is frequently downregulated and functions as a tumor suppressor gene in various cancers [17, 33–36, 40]. Furthermore, the expression levels of miR-223 and miR-375 were not affected by neo-adjuvant chemoradiation therapy [46]. In the current study, we found that abnormal expression of serum miR-25, miR-223, and miR-375 also exists in ESCC patients. The sources of serum miRNAs are relatively unknown. Serum miRNAs can be substances that have been released into the blood as a result of tumor cell apoptosis or disintegration [47]. Valadi et al. [48] showed that mature miRNAs are packaged into exosomes in cells by lipids or lipoproteins and enter the blood through extracellular secretion. Therefore, serum miRNA may also be partly expressed from the active secretion of tissue cells. Our results show that the expression trend of serum miRNA is consistent with that in tumor tissues, suggesting that serum miRNA expression reflect the expression in tumor tissues to a certain extent. Serum samples are convenient to collect, and serum miRNAs are stable enough to serve as optimal tumor biomarkers [49].
Abnormal miRNA expression is associated with tumor formation and development and affects patient prognosis. Dysregulated expressions of miR-223, miR-25 and miR-375 were reported to be associated with poor prognosis in some human cancers [23, 29, 30, 32, 46, 50, 51]. Previous study showed that miR-223 expression in ESCC tissue was inversely associated with the survival, which was correlated to the suppression of the FBXW7 gene functions by high miR-223 expression [32]. Li et al. [30] found that overexpression of miR-223 stimulated nonmetastatic gastric cancer cells migration and invasion and was associated with poor metastasis-free survival. Recent studies showed that low miR-375 expression was associated with worse survival in patients with head and neck squamous carcinoma [23], Barrett’s esophagus and adenocarcinoma [46] or ESCC [36]. MiR-25 can suppress p57 and CDH1 expression, and thereby promote cancer cell migration and invasion [28, 29]. Overexpression of miR-25 in ESCC tissue was associated with a high risk of metastasis [30]. In the present study, we found that abnormal expression of miR-223, miR-25 and miRNA-375 in the serum of ESCC patients are significantly associated with poor prognosis, which were consistent with aforementioned studies. High expression of serum miR-223 and low expression of serum miRNA-375 are the independent markers to predict poor prognosis of ESCC patients. In addition, we combined low expression of serum miR-223 with high expression of serum miRNA-375 for further analysis, and found that ESCC patients without these two risk factors have longer survival. Therefore, serum miR-223 and miR-375 expression may be used as biomarkers to predict survival in ESCC patients. However, studies with adverse conclusions also exist. Karakatsanis et al. [22] revealed that miR-223 expression in liver cancer tissues is not significantly associated with clinical profiles (including metastasis) or survival of patients. Yao et al. [50] suggested that diffuse large B cell lymphoma patients with high miR-223 expression had significantly longer survival time than those with low miR-223 expression. Stamatopoulos et al. [51] reported that reduced miR-223 expression predicted poor prognosis in patients with chronic lymphocytic leukemia. Zhang et al. [52] found that gastric cancer patients with high miR-375 expression had higher risk of recurrence and shorter survival time than those with low expression of miR-375. These inconsistent results may be due to the disparity of tumor tissue sources. Furthermore, mechanisms by which miRNAs function in tumors are extremely complex. One miRNA can control several target genes, whereas several miRNAs can control one target gene. Members of one miRNA cluster can possibly work on target genes. This complex relationship can result in the differential expression between oncogenes and anti-oncogenes in target genes, which eventually affect prognosis. Abnormal expression of miR-223 and miR-375 are associated with the clinicopathological features such as cancer staging and distal metastasis [19, 21]. However, our findings showed no significant association between abnormal expression of serum miR-223 and miRNA-375 and clinicopathological features of ESCC patients. This result may have been caused by different cancers or different mechanisms by which miR-223 and miRNA-375 function.
In summary, we found that ESCC prognosis is associated with the abnormal expression of serum miR-223, miR-25 and miR-375, which could be potential new biomarkers for outcome prediction for ESCC patients. Our findings have provided new insights into studies on the pathogenesis of ESCC and the determination of its prognosis. Suppressed proliferation and induced apoptosis via regulated expression of miRNAs can offer new alternatives in tumor treatment. However, reports on the interaction between miR-223, miR-25 and miR-375 and their corresponding target genes are limited. Functional analysis of these miRNAs will help further reveal the mechanism in the tumorigenesis of ESCC.
References
Jemal A (2011) Global cancer statistics (vol 61, pg 69, 2011). Ca-Cancer J Clin 61(2):134
Portale G, Hagen JA, Peters JH, Chan LS, DeMeester SR, Gandamihardja TA, DeMeester TR (2006) Modern 5-year survival of resectable esophageal adenocarcinoma: single institution experience with 263 patients. J Am Coll Surg 202(4):588–596
Hirst J, Smithers BM, Gotley DC, Thomas J, Barbour A (2011) Defining cure for esophageal cancer: analysis of actual 5-Year survivors following esophagectomy. Ann Surg Oncol 18(6):1766–1774
Lee DH, Kim HR, Kim DK, Park SI, Kim YH (2013) Outcomes of cervical lymph node recurrence in patients with esophageal squamous cell carcinoma after esophagectomy with two-field lymph node dissection. J Thorac Cardiovasc Surg 146(2):365–371
Kosuga T, Shiozaki A, Fujiwara H, Ichikawa D, Okamoto K, Komatsu S, Otsuji E (2011) Treatment outcome and prognosis of patients with lymph node recurrence of thoracic esophageal squamous cell carcinoma after curative resection. World J Surg 35(4):798–804
Hofheinz RD, Al-Batran SE, Ridwelski K, Gorg C, Wehle K, Birth M, Fetscher S, Scheiber H, Lukan N, Lordick F (2010) Population-based patterns of care in the first-line treatment of patients with advanced esophagogastric adenocarcinoma in Germany. Onkologie 33(10):512–518
Zhu Y, Fu L, Chen L, Qin YR, Liu H, Xie F, Zeng T, Dong SS, Li J, Li Y, Dai Y, Xie D, Guan XY (2013) Downregulation of the novel tumor suppressor DIRAS1 predicts poor prognosis in esophageal squamous cell carcinoma. Cancer Res 73(7):2298–2309
Zhai J, Yang X, Zhang Y, Qi Q, Hu J, Wang Q (2013) Reduced expression levels of the death-associated protein kinase and E-cadherin are correlated with the development of esophageal squamous cell carcinoma. Exp Ther Med 5(3):972–976
Ohtsuka M, Yamamoto H, Masuzawa T, Takahashi H, Uemura M, Haraguchi N, Nishimura J, Hata T, Yamasaki M, Miyata H, Takemasa I, Mizushima T, Takiguchi S, Doki Y, Mori M (2013) C4.4A expression is associated with a poor prognosis of esophageal squamous cell carcinoma. Ann Surg Oncol 20(8):2699–2705
Ong CA, Lao-Sirieix P, Fitzgerald RC (2010) Biomarkers in Barrett’s esophagus and esophageal adenocarcinoma: predictors of progression and prognosis. World J Gastroenterol 16(45):5669–5681
Tie J, Fan DM (2011) Big roles of microRNAs in tumorigenesis and tumor development. Histol Histopathol 26(10):1353–1361
Schickel R, Boyerinas B, Park SM, Peter ME (2008) MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. Oncogene 27(45):5959–5974
Fan AC, Goldrick MM, Ho J, Liang Y, Bachireddy P, Felsher DW (2008) A quantitative PCR method to detect blood microRNAs associated with tumorigenesis in transgenic mice. Mol Cancer 7:74
Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120(1):15–20
Kent OA, Mendell JT (2006) A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene 25(46):6188–6196
Vannini I, Fanini F, Fabbri M (2013) MicroRNAs as lung cancer biomarkers and key players in lung carcinogenesis. Clin Biochem 46(10–11):918–925
Faltejskova P, Svoboda M, Srutova K, Mlcochova J, Besse A, Nekvindova J, Radova L, Fabian P, Slaba K, Kiss I, Vyzula R, Slaby O (2012) Identification and functional screening of microRNAs highly deregulated in colorectal cancer. J Cell Mol Med 16(11):2655–2666
Li Q, Zou C, Huang H, Jin J, Han Z, Zhang L, Xiao H, Wei H, Tang Q, Zhang C, Tao J, Wang X, Gao X (2013) MicroRNA-25 functions as a potential tumor suppressor in colon cancer by targeting Smad7. Cancer Lett 335(1):168–174
Kong KL, Kwong DLW, Chan THM, Law SYK, Chen LL, Li Y, Qin YR, Guan XY (2012) MicroRNA-375 inhibits tumour growth and metastasis in oesophageal squamous cell carcinoma through repressing insulin-like growth factor 1 receptor. Gut 61(1):33–42
Yang HS, Gu J, Wang KK, Zhang W, Xing JL, Chen ZN, Ajani JA, Wu XF (2009) MicroRNA expression signatures in Barrett’s esophagus and esophageal adenocarcinoma. Clin Cancer Res 15(18):5744–5752
Gottardo F, Liu CG, Ferracin M, Calin GA, Fassan M, Bassi P, Sevignani C, Byrne D, Negrini M, Pagano F, Gomella LG, Croce CM, Baffa R (2007) Micro-RNA profiling in kidney and bladder cancers. Urol Oncol-Semin Ori 25(5):387–392
Karakatsanis A, Papaconstantinou I, Gazouli M, Lyberopoulou A, Polymeneas G, Voros D (2013) Expression of microRNAs, miR-21, miR-31, miR-122, miR-145, miR-146a, miR-200c, miR-221, miR-222, and miR-223 in patients with hepatocellular carcinoma or intrahepatic cholangiocarcinoma and its prognostic significance. Mol Carcinog 52(4):297–303
Harris T, Jimenez L, Kawachi N, Fan JB, Chen J, Belbin T, Ramnauth A, Loudig O, Keller CE, Smith R, Prystowsky MB, Schlecht NF, Segall JE, Childs G (2012) Low-level expression of miR-375 correlates with poor outcome and metastasis while altering the invasive properties of head and neck squamous cell carcinomas. Am J Pathol 180(3):917–928
Liu R, Zhang CN, Hu ZB, Li G, Wang C, Yang CH, Huang DZ, Chen X, Zhang HY, Zhuang R, Deng T, Liu H, Yin JJ, Wang SF, Zen K, Ba Y, Zhang CY (2011) A five-microRNA signature identified from genome-wide serum microRNA expression profiling serves as a fingerprint for gastric cancer diagnosis. Eur J Cancer 47(5):784–791
Brase JC, Johannes M, Schlomm T, Falth M, Haese A, Steuber T, Beissbarth T, Kuner R, Sultmann H (2011) Circulating miRNAs are correlated with tumor progression in prostate cancer. Int J Cancer 128(3):608–616
Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A, Lin DW, Urban N, Drescher CW, Knudsen BS, Stirewalt DL, Gentleman R, Vessella RL, Nelson PS, Martin DB, Tewari M (2008) Circulating microRNAs as stable blood-based markers for cancer detection. P Natl Acad Sci USA 105(30):10513–10518
Zen K, Zhang CY (2012) Circulating MicroRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev 32(2):326–348
Kim YK, Yu J, Han TS, Park SY, Namkoong B, Kim DH, Hur K, Yoo MW, Lee HJ, Yang HK, Kim VN (2009) Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res 37(5):1672–1681
Chen X, Hu Z, Wang W, Ba Y, Ma L, Zhang C, Wang C, Ren Z, Zhao Y, Wu S, Zhuang R, Zhang Y, Hu H, Liu C, Xu L, Wang J, Shen H, Zhang J, Zen K, Zhang CY (2012) Identification of ten serum microRNAs from a genome-wide serum microRNA expression profile as novel noninvasive biomarkers for nonsmall cell lung cancer diagnosis. Int J Cancer 130(7):1620–1628
Li X, Zhang Y, Zhang H, Liu X, Gong T, Li M, Sun L, Ji G, Shi Y, Han Z, Han S, Nie Y, Chen X, Zhao Q, Ding J, Wu K, Daiming F (2011) miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res 9(7):824–833
Li BS, Zhao YL, Guo G, Li W, Zhu ED, Luo X, Mao XH, Zou QM, Yu PW, Zuo QF, Li N, Tang B, Liu KY, Xiao B (2012) Plasma microRNAs, miR-223, miR-21 and miR-218, as novel potential biomarkers for gastric cancer detection. PLoS One 7(7):e41629
Kurashige J, Watanabe M, Iwatsuki M, Kinoshita K, Saito S, Hiyoshi Y, Kamohara H, Baba Y, Mimori K, Baba H (2012) Overexpression of microRNA-223 regulates the ubiquitin ligase FBXW7 in oesophageal squamous cell carcinoma. Brit J Cancer 106(1):182–188
Tsukamoto Y, Nakada C, Noguchi T, Tanigawa M, Lam TN, Uchida T, Hijiya N, Matsuura K, Fujioka T, Seto M, Moriyama M (2010) MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3 zeta. Cancer Res 70(6):2339–2349
Dai X, Chiang Y, Wang Z, Song Y, Lu C, Gao P, Xu H (2012) Expression levels of microRNA-375 in colorectal carcinoma. Mol Med Rep 5(5):1299–1304
Chang Y, Yan W, He X, Zhang L, Li C, Huang H, Nace G, Geller DA, Lin J, Tsung A (2012) miR-375 inhibits autophagy and reduces viability of hepatocellular carcinoma cells under hypoxic conditions. Gastroenterol 143(1):177–187
Li J, Li X, Li Y, Yang H, Wang L, Qin Y, Liu H, Fu L, Guan XY (2013) Cell-specific detection of miR-375 downregulation for predicting the prognosis of esophageal squamous cell carcinoma by miRNA in situ hybridization. PLoS One 8(1):e53582
Zhang HY, Zuo Z, Lu X, Wang L, Wang HY, Zhu ZL (2012) MiR-25 regulates apoptosis by targeting Bim in human ovarian cancer. Oncol Rep 27(2):594–598
Esposito F, Tornincasa M, Pallante P, Federico A, Borbone E, Pierantoni GM, Fusco A (2012) Down-regulation of the miR-25 and miR-30d contributes to the development of anaplastic thyroid carcinoma targeting the polycomb protein EZH2. J Clin Endocrinol Metab 97(5):E710–E718
Zhou K, Yi S, Yu Z, Li Z, Wang Y, Zou D, Qi J, Zhao Y, Qiu L (2012) MicroRNA-223 expression is uniformly down-regulated in B cell lymphoproliferative disorders and is associated with poor survival in patients with chronic lymphocytic leukemia. Leuk Lymphoma 53(6):1155–1161
Kinoshita T, Hanazawa T, Nohata N, Okamoto Y, Seki N (2012) The functional significance of microRNA-375 in human squamous cell carcinoma: aberrant expression and effects on cancer pathways. J Hum Genet 57(9):556–563
Kurashige J, Kamohara H, Watanabe M, Tanaka Y, Kinoshita K, Saito S, Hiyoshi Y, Iwatsuki M, Baba Y, Baba H (2012) Serum microRNA-21 is a novel biomarker in patients with esophageal squamous cell carcinoma. J Surg Oncol 106(2):188–192
Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM (2007) MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA 297(17):1901–1908
Laios A, O’Toole S, Flavin R, Martin C, Kelly L, Ring M, Finn SP, Barrett C, Loda M, Gleeson N, D’Arcy T, McGuinness E, Sheils O, Sheppard B, Leary JO (2008) Potential role of miR-9 and miR-223 in recurrent ovarian cancer. Mol Cancer 7:35
Belair C, Darfeuille F, Staedel C (2009) Helicobacter pylori and gastric cancer: possible role of microRNAs in this intimate relationship. Clin Microbiol Infec 15(9):806–812
Liu XG, Zhu WY, Huang YY, Ma LN, Zhou SQ, Wang YK, Zeng F, Zhou JH, Zhang YK (2012) High expression of serum miR-21 and tumor miR-200c associated with poor prognosis in patients with lung cancer. Med Oncol 29(2):618–626
Mathe EA, Nguyen GH, Bowman ED, Zhao YQ, Budhu A, Schetter AJ, Braun R, Reimers M, Kumamoto K, Hughes D, Altorki NK, Casson AG, Liu CG, Wang XW, Yanaihara N, Hagiwara N, Dannenberg AJ, Miyashita M, Croce CM, Harris CC (2009) MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res 15(19):6192–6200
Chin LJ, Slack FJ (2008) A truth serum for cancer—microRNAs have major potential as cancer biomarkers. Cell Res 18(10):983–984
Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9(6):654–659
Chen X, Ba Y, Ma LJ, Cai X, Yin Y, Wang KH, Guo JG, Zhang YJ, Chen JN, Guo X, Li QB, Li XY, Wang WJ, Zhang Y, Wang J, Jiang XY, Xiang Y, Xu C, Zheng PP, Zhang JB, Li RQ, Zhang HJ, Shang XB, Gong T, Ning G, Wang J, Zen K, Zhang JF, Zhang CY (2008) Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res 18(10):997–1006
Yao XX, Wang JF, Wang YH, Gao N (2012) Expression of microRNA-223 and its clinicopathologic correlation in diffuse large B-cell lymphoma. Zhonghua Bing Li Xue Za Zhi 41(6):366–370
Stamatopoulos B, Meuleman N, Haibe-Kains B, Saussoy P, Van Den Neste E, Michaux L, Heimann P, Martiat P, Bron D, Lagneaux L (2009) microRNA-29c and microRNA-223 down-regulation has in vivo significance in chronic lymphocytic leukemia and improves disease risk stratification. Blood 113(21):5237–5245
Zhang X, Yan Z, Zhang J, Gong L, Li W, Cui J, Liu Y, Gao Z, Li J, Shen L, Lu Y (2011) Combination of hsa-miR-375 and hsa-miR-142-5p as a predictor for recurrence risk in gastric cancer patients following surgical resection. Ann Oncol 22(10):2257–2266
Acknowledgments
We thank Yuanjirong and Haixian Lu for reading this paper and for their valuable suggestions.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2013_2970_MOESM1_ESM.tif
Fig. S1. Comparison analysis of the change pattern in 20 matched tissue and serum samples. a miR-25. b miR-223. c miR-375. (TIFF 95 kb)
Rights and permissions
About this article
Cite this article
Wu, C., Li, M., Hu, C. et al. Clinical significance of serum miR-223, miR-25 and miR-375 in patients with esophageal squamous cell carcinoma. Mol Biol Rep 41, 1257–1266 (2014). https://doi.org/10.1007/s11033-013-2970-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-013-2970-z