Abstract
Prior to the development of zinc-finger nuclease technology, genetic manipulation by gene targeting achieved limited success in mammals, with the exception of mice and rat. Although ZFNs demonstrated highly effective gene targeted disruption in various model organisms, the activity of ZFNs in large domestic animals may be very low, and the probability of identifying ZFN-mediated positive targeted disruption events is small. In this paper, we used the context-dependent assembly method to synthesize two pairs of ZFNs targeted to the sheep MSTN gene. We verified the activity of these ZFNs using an mRFP-MBS-eGFP dual-fluorescence reporter system in HEK293T cells and, according to the expression level of eGFP, we obtained a pair of ZFNs that can recognize and cut the targeted MSTN site in the reporter vector. The activity of ZFN was increased by cold stimulation at 30 °C and by mutant the wildtype FokI in ZFN with its counterpart Sharkeys. Finally, the ZF-Sharkeys and reporter vector were cotransfected into sheep fetal fibroblasts and two MSTN mutant cell lines, identified by flow cytometry and sequencing, were obtained.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Targeted gene modification is a powerful technique for investigating gene function, producing models of human disease and breeding animals with desired traits. It also holds much promise for the therapy of human genetic diseases. Traditional gene targeting techniques in embryonic stem (ES) cells, followed by germline chimera production, have been used to generate a vast number of gene-targeted mice [1, 2]. However, this technology is difficult to transfer from mice to large animals, such as swine, sheep and cattle, because the shortage of ES cells derived from domestic animals, and the inherently low efficiency of spontaneous homologous recombination in mammalian somatic cell. It has been demonstrated that the introduction of double-strand breaks (DSBs) in chromosomes can significantly stimulate the efficiency of homologous recombination [3]. Recently, gene knockout can be achieved by using artificial zinc-finger nucleases (ZFNs) to introduce a DSBs at the interest site of chromosomal DNA sequences, which can be be spontaneously repaired by non-homologous-end-joining (NHEJ), in the absence of donor DNA. The NHEJ repair pathway may result in micro-insertions or micro-deletions at targeted region, leading to loss-of-function gene. As a robust gene editing tool, ZFNs successfully modified genomes in various organisms, including Drosophila [4], Arabidopsis thaliana [5], zebrafish [6, 7], mouse [8], silkworm (Bombyx mori) [9], frog [10], rat [11], human [12], pig [13], rabbit [14], cattle [15] and yellow catfish [16], and the targeted modification frequency was up to 50 % without drug selection.
Myostatin (MSTN) is an important negative regulator of muscle growth and belongs to the transforming growth factor-β family [17]. It has been reported that MSTN mutation enhanced muscle growth or “double muscle”, which happens naturally in cattle, dogs, sheep and humans [18]. Also, it has been demonstrated that homozygous knockout of MSTN in mouse by conventional homologous recombination results in increased muscle mass [19], which has prominent economic value for meat production. In livestock, however, MSTN knockout has not been achieved. Dong et al. [16] have performed targeted disruption of MSTN in yellow catfish using ZFNs; however, no studies reported about the suitability of ZFNs for the targeted modification in the sheep genome. In this study, we used the CoDA (context-dependent assembly) method [20] to design and assemble two pairs of ZFNs, which target against the sheep MSTN locus within exon 1 to disrupt the MSTN for new sheep breed generation purpose.
Materials and methods
Vector construction
ZFN expression vectors
According to the CoDA method, the ZF binding sites were identified with the tool provided by the web based software (http://www.zincfingers.org/software-tools.htm). Only two potential target sites were screened within MSTN gene exons, and so corresponding two pairs of zinc-finger sequences (Table 1) were synthesized and cloned into pST1374, respectively. To increase the activity of synthesized ZFNs, the wildtype FokI in pST1374 were replaced with Sharkey [21]. Four pairs of ZFN expression vectors, termed ZFN1 (pZFN-1L/pZF-1R), ZFN2 (pZFN-2L/pZFN-2R), ZFS1 (pZFS-1L/pZFS-1R) and ZFS2 (pZFS-2L/pZFS-2R), were constructed.
Reporter vectors
Two pairs of oligonucleotides (MBS-F and MBS-R in Table 2) harboring the MSTN ZFN target sequences were synthesized and annealed to cloned into reporter vector pmRFP-eGFP between BamHI/NotI sites, and the obtained reporter vector was named pRGS. In pRGS, the eGFP open reading frame (ORF) was disrupted by an inserted stop codon and a MSTN ZFN target site which divided the eGFP ORF into two segments. To facilitate the homologous recombination between the two segments they were set a 200-bp homologous region (supplemental Fig. 1).
Cell culture and transfection
Human embryonic kidney 293T (HEK293T) cells
HEK293T cells were cultured in Dulbecco’s modified eagle’s medium (DMEM; Sigma) supplemented with 100 U/ml penicillin, 100 g/ml streptomycin and 10 % (v/v) fetal bovine serum (FBS; Hyclone, Logan, State, USA), at 37 °C in an atmosphere of 5 % CO2. About 1 × 104 HEK293T cells were seeded into each well of a 24-well plate in 0.5 ml of culture medium and incubated for 24 h before transfection. Culture medium was replaced carefully 1 h prior to transfection and transfection mix was prepared in a 1.5 ml tube as follows: 1 μg DNA in 47 μl Opti-MEM (Sigma) and 2.5 μl Lipofectamine 2000 (Invitrogen) in 47.5 μl Opti-MEM. The solution was mixed carefully, incubated for 25 min at room temperature, and then added dropwise to cells. Cells were then incubated at 37 °C in an atmosphere of 5 % CO2.
Sheep fetal fibroblasts
Sheep fetal fibroblasts were obtained from an adult sheep as described by Hu et al. [22]. The fibroblasts were maintained in DMEM supplemented with 100 U/ml penicillin, 100 g/ml streptomycin and 15 % (v/v) FBS (Hyclone), at 37 °C in an atmosphere of 5 % CO2. About 3 × 106 sheep fetal fibroblasts were seeded in 100-mm dishes in 10 ml of culture medium and incubated for 48 h before transfection. Cells were digested with 0.05 % trypsin–EDTA and resuspended in 10 ml Opti-MEM, then quantified with a hemocytometer. Cells were resuspended in transfection medium [75 % cytosalts (120 mM KCl, 0.15 mM CaCl2, 10 mM K2HPO4; pH 7.6, 5 mM MgCl2) [23], 25 % Opti-MEM] and the cell number was adjusted to 1 × 106 cells/ml. Electroporations were performed in 4-mm gap cuvettes containing 400 μl transfection medium, 1 × 106 cells/ml and 6 μg plasmid with two 1-ms pulses of 500 V, administered through an ECM 2001 electro cell manipulator (BTX, Holliston, MA, USA).
ZFN activity assay
To assay the activity of synthesized ZFNs, HEK293T cells were co-transfected with pRGS and each pair of synthesized ZFNs, respectively, the vector ratio were all 4:3:3. And HEK293 cells transfected with reporter pRGS only were set as control. To test the effect of culture temperature on ZFN activity, the cells were divided into two equal parts and incubated for 3 days at 37 °C, or for 1 day at 37 °C followed by 2 days at 30 °C, respectively. Cells were observed 24, 48 and 72 h after transfection under a fluorescence microscope. Seventy-two hours after transfection, 30,000 cells were counted and the number of RFP express (Rex)-positive cells and eGFP express (GEx)-positive cells evaluated by flow cytometry. The percentage of REx-positive cells represents the efficiency of transfection, and the percentage of GEx-positive cells reflects the activity of ZFNs.
ZFN-targeted MSTN in sheep fetal fibroblasts
Sheep fetal fibroblasts were transfected with the reporter pRGS only, co-transfected with pRGS, pZFN-1L and pZFN-1R (4:3:3), or co-transfected with pRGS, pZFS-1L and pZFS-1R (4:3:3). The transfected cells were incubated for 1 day at 37 °C followed by 2 days at 30 °C. Three days after transfection, the adherent cells were trypsinized and resuspended in DMEM (Sigma) supplemented with 100 U/ml penicillin, 100 g/ml streptomycin and 15 % (v/v) FBS. Single-cell suspensions were sorted using a FACSAria II (BD Biosciences). To collect cells that contain nuclease-induced mutations, cells with strong eGFP signals were sorted. Untransfected cells and cells transfected with reporters alone were used as controls.
T7E1 assay
The T7E1 assay was performed as previously described [8]. Briefly, genomic DNA was isolated using the DNeasy Blood and Tissue Kit (Qiagen) according to the manufacturer’s instructions. PCR reactions (50 μl) were carried out using one unit high fidelity Pfu DNA Polymerase (GenScript) in the manufacturer-supplied buffer with 0.4 mM dNTPs and 10 μM primers (MSTN-F and MSTN-R in Table 2). This amplification should produce a 342 bp DNA fragment with the ZFN target site at its center. About 100 ng genomic DNA was used as the template in each PCR reaction. PCR reaction program: denature at 95 °C for 4 min followed by 35 cycles consisting of 94 °C for 30 s, 58 °C for 30 s and 72 °C for 30 s; 72 °C for 10 min final extension. The amplicons were denatured by heating and reannealed to form heteroduplex DNA, which was treated with 1 unit of T7 endonuclease 1(T7E1, New England Biolabs) for 1 h at 37 °C and then analyzed by agarose gel electrophoresis.
Analysis of clones derived from single cells
The eGFP-positive FACSAria-sorted cells were plated at a density of 500 cells per 100-mm dish. The culture medium was changed every 3 days. After culture for about 14 days, the cell clones were picked out into 24-well culture dishes to expand the clones. After cells were nearly confluent, they were harvested by trypsin digestion and then divided into two aliquots. One aliquot was expanded to confluence in two wells of a 24-well culture dish and cryopreserved. The other aliquot was used to identify targeting events. Cells were lysed with embryo lysis buffer according to Dai et al [24] and the lysate was used as template for PCR amplification of the targeted locus. The amplified fragment was purified, ligated into pMD18 T and sequenced.
Results
To knock out the MSTN gene in sheep, we designed and synthesized two pairs of ZFNs according to the CoDA method. Both of the ZFN-targeted sites were located in exon 1 of MSTN (Fig. 1). To determine ZFN activity and to enrich ZFN-targeted cells, we constructed a surrogate reporter in which the ZFN target site cassette disrupts the ORF of an eGFP reporter gene. If the targeted site is cut by the ZFN, a DSB will generated. This will promote homologous recombination between the two repeat regions of eGFP gene and restore the expression of eGFP, which can be detected by fluorescence microscopy or flow cytometry [25].
The activity of synthesized ZFNs was determined using the reporter system in HEK293T cells. Seventy-two hours after transfection, a similar number of cells expressing mRFP were detected in all groups of treated cells. Some of the mRFP expressing cells showed simultaneously green fluorescence when transfected with the reporter plasmid and plasmids encoding ZFN1, but few cells expressed eGFP when transfected with the reporter only or when co-transfected with the reporter plasmid and plasmids encoding ZFN2 (data was not shown). These results suggested that ZFN1 can recognize and cut the MSTN ZFN target site on pRGS reporter vector more efficiently than ZFN2.
To test the effect of culture temperature on ZFN activity, the cells were divided into 2 equal parts and incubated for 3 days at 37 °C, or for 1 day at 37 °C followed by 2 days at 30 °C, respectively. It was observed that the cells with eGFP expression cultured 1 day at 37 °C followed by another 2 days at 30 °C were a little more than the cells cultured all 3 days at 37 °C but there was no statistically difference (as shown in supplemental Fig. 2).
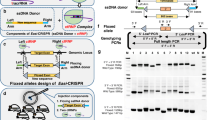
To enhance the efficiency of the ZFNs activity, we changed the FokI nuclease domain in ZFN1 and ZFN2 with the Sharkey cleavage domain, according to the study of Guo et al. [21]. The resulting ZFN mutants were termed ZFS1 and ZFS2. The activities of ZFS1 and ZFS2 were determined by cotransfection with the reporter system in HEK293T cells. As shown in Fig. 2, flow cytometry indicated that the efficiency of transfection was about 55 % and eGFP was expressed in approximately 15.06 % of cells cotransfected with the reporter plasmid and plasmids encoding ZFN1, in 30.05 % of cells cotransfected with the reporter and ZFS1, in 8.65 % of cells cotransfected with the reporter and ZFS2 and in only 4.14 % of cells transfected with the reporter and ZFN2. These results indicated that the ZFN mutants ZFS1 and ZFS2 can cut the MSTN ZFN target site located in the reporter vector with higher efficiency than their wild-type counterparts.
Efficiency of RFP and eGFP expressions in HEK293T cells cotransfected with a reporter plasmid and plasmids encoding wild-type ZFNs or mutant ZFSs (ZF-Sharkey). HEK293T cells were subjected to flow cytometry 72 h after transfection. Untransfected cells and cells transfected with the surrogate reporter plasmid were used as control and cells transfected with the reporter and plasmids encoding different ZFN paris were analyzed, respectively
Based on the above findings, we used the ZFS1 plasmids combined with the reporter to disrupt the MSTN gene in sheep fetal fibroblasts. The treated cells were analyzed and sorted using a FACSAria II, and approximately 30,000 eGFP-expressing cells from three 100-mm dishes were collected. Only 1.7 % of cells expressed eGFP were sorted (Fig 3). From the eGFP-expressing cells, approximately 10,000 cells were used to isolate genomic DNA for the T7E1 assay, and the remaining cells were plated at a density of 500 cells per 100-mm dish. The T7E1 assay (Fig 4e) displayed three bands but two were very weak in the sorted cells. Only one band was produced from unsorted cells, which suggested that genome-modified cells were enriched by the dual fluorescent reporter vector. After screening 80 clones by sequencing, 2 cell clones with the disruption of MSTN were detected. The efficiency of ZFS1 targeting was approximately 2.5 %. As shown in Fig. 4a–c, the sequencing ideograms show double peaks after the targeting site. A 3 bp deletion and a 5 bp deletion in the cutting site were detected by analysis of the sheep MSTN gene sequence (no.DQ530260) in the two clones, respectively (Fig. 4d).
Representative results of sequencing traces of a wild-type (a) and two mutant (b, c) clones. (d) Genetic lesions identified in mutant cells derived from individual clones. The blue box represents the ZFN cutting site; black dashes indicate the positions of deletions in the genomic sequences. (e) ZFN-driven mutations detected by the T7E1 assay. The genomic DNA was isolated from the flow-cytometrically sorted cells and unsorted cells, and subjected to the T7E1 assay. The sorted are the MSTN gene-modified cells, which were enriched in the RFP+ GFP+ cells by flow cytometry as compared to unsorted cells. The bands indicated by the arrows stand for the amplicon cut by T7E1, ‘unsorted’ lane has only one band, ‘sorted’ lanes have three bands
Discussion
Gene targeting is a powerful technique for disrupting an endogenous gene, inserting an exogenous gene fragment into a specific locus, or subtly modifying a gene. However, owing to the inherently low efficiency of naturally occurring homologous recombination in mammalian somatic cell, traditional gene targeting cannot be used extensively. It has been reported that the naturally occurring MSTN mutant can result in excess muscle development called “double muscle”[26]. MSTN has previously been artificially mutated in vitro in bovine and ovine cells, but the efficiency was too low to select a targeted cell clone with expected growth performance [27, 28]. Recently, ZFNs, which can be artificially designed, have shown great potential for targeted modification of specific loci in zebrafish, mouse, rat and even domestic animals such as swine and cattle. With the help of ZFNs, Dong et al. [16] successfully gained some yellow catfish with MSTN disruption [16]. However, most of these ZFNs were supplied by Sigma-Aldrich [7] with a extremely high cost, and only a few were synthesized with the modular method [29], OPEN [30], or by the recently developed CoDA method [20]. The main bottlenecks limit the application of ZFNs designed by the OPEN and CoDA methods are relatively impotent specificity or affinity compared with Sigma-Aldrich supplied ZFNs and the lack of a system to enrich or select for targeted cells. And even with the ZFNs supplied by Sigma-Aldrich, some suitable systems may be helpful to enrich targeted cells. By removing Gal+ cells with magnetic bead-based method, 99 % of enriched cells were believed to be Gal− analyzed by FACS [13]. The ZFNs synthesized by us using the CoDA method showed low cleavage efficiency, as tested by recovery of eGFP expression and the T7E1 test. T7E1 test was widely used in lots of studies to detect the activity of ZFNs, but it was tedious and not so sensitive.
Firstly, we constructed a surrogate reporter system to detect the activity of ZFNs. The surrogate reporter vector was constructed based on the principle reported by Kim et al. [24]. In our reporter plasmid, instead of being located in the intergenic region between mRFP and eGFP, the target site and a stop codon were inserted in the eGFP ORF, which divided the ORF into two segments. Two direct repeats of 275 bp were introduced on either side of the target site according to Perez-Pinera et al. [31]. Thus, if the target site was recognized and cleaved by the corresponding ZFN, homologous recombination can take place leading to recovery of the eGFP ORF. The expression of eGFP can be detected directly by fluorescence microscope or FACS. Perez et al. [32] reported that when the repeat sequences separated by the target site was 220 bp, up to 50 % of the extra-chromosomal plasmid could be modified, but only 0.5–3.5 % of sites on the chromosome were recovered. Less than 20 % of eGFP loci disrupted by the MSTN ZFN target site in pRGS were recovered in our study, which suggested that less chromosomal homologous recombination may occur. Therefore, two methods to enhance the activity of synthesized ZFN and one technique to enrich the targeted cells were employed.
To increase the efficiency of the ZFN, we reduced the temperature of the cell culture according to Doyon et al. [33]. When transfected cells were incubated for 1 day at 37 °C followed by 2 days at 30 °C, there were slightly more cells expressing green fluorescence compared with the other two culture conditions, but these differences were not statistically significant. It has been found that the specificity and affinity of ZFNs can be enhanced by mutation of the FokI nuclease domain. To improve the cleavage efficiency of ZFNs further, the Sharkey domain, described by Guo et al. [25], was chosen to replace the ZFN FokI motif. As reported by Guo et al., Sharkey was 15-fold more efficient than the wild-type FokI domain on a diverse panel of cleavage sites. Our results showed that the cleavage activity of Sharkey was twice than that of the wild-type FokI nuclease.
To select more ZFN targeted cells, the reporter vector used for ZFN activity assay was utilized to enrich the targeted cells. In the surrogate vector designed by Kim [24], which was taken by other researchers in enriching TALEN targeted cells [34], the target site was located between DNA sequences encoding mRFP and eGFP. As described by Kim, only one-third of the reporter plasmids can be fused in-frame to recover eGFP expression owing to the random occurrence of non-homologous end joining. Therefore, many out-of-frame cells may be missed. It has been reported that when DSBs are introduced, the frequency of homologous recombination will increase more than 1,000 times, which is comparable with non-homologous end joining. Liang et al. reported that when a DSB was introduced into one of the two direct repeats, homologous repair can account for 30–50 % of observed repair events, which suggested that homologous recombination is a major DSB repair pathway in mammalian cells [35]. A similar reporter system based on homologous recombination has been used to detect the activity of ZFNs [36]. When reporter plasmids were cotransfected with corresponding ZFNs, the targeted cells were enriched 59 times compared with cells treated with ZFNs only.
In this study, one ZFN induced 3-bp deletion in sheep MSTN gene should not alter the open reading frame, just leads to an amino acid delete. The second mutant, a 5-bp deletion, which leads to frameshift mutation, thus it would likely to generate gene inactivation. Furthermore, we will continue to study whether the delete mutation and frameshift mutation make MSTN inactivation to obtain expected growth performance. To date, ZFNs have been successfully used to disrupt α-1, 3-galactosyltransferase (GGTA1) and PPARγ in pig and β-lactoglobulin (BLG) in cattle and they showed great potential for the targeted modification of domestic animals. For the first time, we report the targeted disruption of MSTN in sheep using ZFNs. With the help of our surrogate reporter system, a targeted disruption efficiency was up to 2.5 %, which is more efficient compared with traditional gene targeted disruption of MSTN in sheep [28]. We propose that our reporter system, together with Sharkey, may facilitate the use of ZFNs in the targeted modification of mammalian genes.
References
Thomas KR, Folger KR, Capecchi MR (1986) High frequency targeting of genes to specific sites in the mammalian genome. Cell 44(3):419–428
Robertson EJ (1991) Using embryonic stem cells to introduce mutations into the mouse germ line. Biol Reprod 44(2):238–245
Rouet P, Smih F, Jasin M (1994) Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proc Natl Acad Sci 91(13):6064–6068
Bibikova M, Golic M, Golic KG, Carroll D (2002) Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161(3):1169–1175
Lloyd A, Plaisier CL, Carroll D, Drews GN (2005) Targeted mutagenesis using zinc-finger nucleases in Arabidopsis. Proc Natl Acad Sci USA 102(6):2232–2237
Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA (2008) Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol 26(6):695–701
Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ (2008) Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol 26(6):702–708
Meyer M, de Angelis MH, Wurst W, Kühn R (2010) Gene targeting by homologous recombination in mouse zygotes mediated by zinc-finger nucleases. Proc Natl Acad Sci 107(34):15022–15026
Takasu Y, Kobayashi I, Beumer K, Uchino K, Sezutsu H, Sajwan S, Carroll D, Tamura T, Zurovec M (2010) Targeted mutagenesis in the silkworm Bombyx mori using zinc finger nuclease mRNA injection. Insect Biochem Mol Biol 40(10):759–765
Young JJ, Cherone JM, Doyon Y, Ankoudinova I, Faraji FM, Lee AH, Ngo C, Guschin DY, Paschon DE, Miller JC (2011) Efficient targeted gene disruption in the soma and germ line of the frog Xenopus tropicalis using engineered zinc-finger nucleases. Proc Natl Acad Sci 108(17):7052–7057
Chu X, Zhang Z, Yabut J, Horwitz S, Levorse J, XQ Li, Zhu L, Lederman H, Ortiga R, Strauss J (2012) Characterization of multidrug resistance 1a/P-glycoprotein knockout rats generated by zinc finger nucleases. Mol Pharmacol 81(2):220–227
Moehle EA, Rock JM, Lee YL, Jouvenot Y, DeKelver RC, Gregory PD, Urnov FD, Holmes MC (2007) Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc Natl Acad Sci 104(9):3055–3060
Hauschild J, Petersen B, Santiago Y, Queisser AL, Carnwath JW, Lucas-Hahn A, Zhang L, Meng X, Gregory PD, Schwinzer R (2011) Efficient generation of a biallelic knockout in pigs using zinc-finger nucleases. Proc Natl Acad Sci 108(29):12013–12017
Flisikowska T, Thorey IS, Offner S, Ros F, Lifke V, Zeitler B, Rottmann O, Vincent A, Zhang L, Jenkins S (2011) Efficient immunoglobulin gene disruption and targeted replacement in rabbit using zinc finger nucleases. PLoS ONE 6(6):e21045
Yu S, Luo J, Song Z, Ding F, Dai Y, Li N (2011) Highly efficient modification of beta-lactoglobulin (BLG) gene via zinc-finger nucleases in cattle. Cell Res 21(11):1638–1640
Dong Z, Ge J, Li K, Xu Z, Liang D, Li J, Li J, Jia W, Li Y, Dong X, Cao S, Wang X, Pan J, Zhao Q (2011) Heritable targeted inactivation of myostatin gene in yellow catfish (Pelteobagrus fulvidraco) using engineered zinc finger nucleases. PLoS ONE 6(12):e28897
McPherron AC, Lawler AM, Lee SJ (1997) Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature 387(6628):83–90
Stinckens A, Georges M, Buys N (2011) Mutations in the Myostatin gene leading to hypermuscularity in mammals: indications for a similar mechanism in fish? Anim Genet 42(3):229–234
Chelh I, Picard B, Hocquette J, Cassar-Malek I (2011) Myostatin inactivation induces a similar muscle molecular signature in double-muscled cattle as in mice. Animal 5(02):278–286
Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y (2010) Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat Methods 8(1):67–69
Guo J, Gaj T, Barbas CF (2010) Directed evolution of an enhanced and highly efficient FokI cleavage domain for zinc finger nucleases. J Mol Biol 400(1):96–107
Hu LY, Cui CC, Song YJ, Wang XG, Jin YP, Wang AH, Zhang Y (2012) An alternative method for cDNA cloning from surrogate eukaryotic cells transfected with the corresponding genomic DNA. Biotechnol Lett 34(7):1251–1255
Van den Hoff M, Moorman A, Lamers WH (1992) Electroporation in ‘intracellular’ buffer increases cell survival. Nucleic Acids Res 20(11):2902
Dai Y, Vaught TD, Boone J, Chen SH, Phelps CJ, Ball S, Monahan JA, Jobst PM, McCreath KJ, Lamborn AE (2002) Targeted disruption of the alpha1, 3-galactosyltransferase gene in cloned pigs. Nat Biotechnol 20(3):251–255
Kim H, Um E, Cho SR, Jung C, Kim H, Kim JS (2011) Surrogate reporters for enrichment of cells with nuclease-induced mutations. Nat Methods 8(11):941–943
McPherron AC, Lee SJ (1997) Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci 94(23):12457–12461
Liu Y, Hua S, Lan J, Song Y, He Y, Quan F, Zhang Y (2010) Site-directed mutagenesis of MSTN gene by gene targeting in Qinchuan cattle. Sheng wu gong cheng xue bao 26(3):410–416 Chinese Journal of Biotechnology
Zheng YL, Ma HM, Zheng YM, Wang YS, Zhang BW, He XY, He XN, Liu J, Zhang Y (2012) Site-directed mutagenesis of the myostatin gene in ovine fetal myoblast cells in vitro. Res Vet Sci 93(2):763–769
Kim JS, Lee HJ, Carroll D (2010) Genome editing with modularly assembled zinc-finger nucleases. Nat Methods 7(2):91
Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA (2008) Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell 31(2):294–301
Perez-Pinera P, Ousterout DG, Brown MT, Gersbach CA (2012) Gene targeting to the ROSA26 locus directed by engineered zinc finger nucleases. Nucleic Acids Res 40(8):3741–3752
Perez C, Guyot V, Cabaniols JP, Gouble A, Micheaux B, Smith J, Leduc S, Paques F, Duchateau P (2005) Factors affecting double-strand break-induced homologous recombination in mammalian cells. Biotechniques 39(1):109–115
Doyon Y, Choi VM, Xia DF, Vo TD, Gregory PD, Holmes MC (2010) Transient cold shock enhances zinc-finger nuclease-mediated gene disruption. Nat Methods 7(6):459–460
Davies B, Davies G, Preece C, Puliyadi R, Szumska D, Bhattacharya S (2013) Site specific mutation of the Zic2 locus by microinjection of TALEN mRNA in mouse CD1, C3H and C57BL/6J oocytes. PLoS ONE 8(3):e60216
Liang F, Han M, Romanienko PJ, Jasin M (1998) Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc Natl Acad Sci 95(9):5172–5177
Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC (2005) Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435(7042):646–651
Acknowledgments
This study was supported by a grant from the genetically modified organisms breeding major projects of China (2013XZ08008-003).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, C., Wang, L., Ren, G. et al. Targeted disruption of the sheep MSTN gene by engineered zinc-finger nucleases. Mol Biol Rep 41, 209–215 (2014). https://doi.org/10.1007/s11033-013-2853-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-013-2853-3