Abstract
Salvia miltiorrhiza Bunge is a well-known material of traditional Chinese medicine. Hydrophilic phenolic acids, such as rosmarinic acid and salvianolic acid B, are a group of pharmaceutically important compounds in S. miltiorrhiza. The biosynthesis of rosmarinic acid requires the coordination of the phenylpropanoid pathway and the tyrosine-derived pathway. Phenylalanine ammonia-lyase (PAL) is the first key enzyme of the phenylpropanoid pathway. Systematic analysis of the SmPAL gene family has not been carried out. We report here the identification of three SmPALs through searching the recently obtained working draft of the S. miltiorrhiza genome and full-length cDNA cloning. Bioinformatic and phylogenetic analyses showed that SmPAL1 and SmPAL3 clustered in a sub-clade of dicot PALs, whereas SmPAL2 fell into the other one. Some important cis-elements were conserved in three SmPAL promoters, whereas the others were not. SmPAL1 and SmPAL3 were highly expressed in roots and leaves of S. miltiorrhiza, but SmPAL2 were predominately expressed in stems and flowers. It indicates that SmPAL1 and SmPAL3 function redundantly in rosmarinic acid biosynthesis. All SmPALs were induced in roots treated with PEG and MeJA, but the time and degree of responses were different, suggesting the complexity of SmPAL-associated metabolic network in S. miltiorrhiza. This is the first comprehensive study dedicated to SmPAL gene family characterization. The results provide a basis for elucidating the role of SmPAL genes in the biosynthesis of bioactive compounds.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
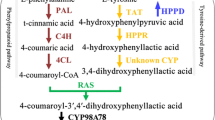
Salvia miltiorrhiza Bunge is a well-known material of various traditional Chinese medicines (TCMs) that have been widely used for treating dysmenorrhoea, amenorrhoea, and cardiovascular diseases [1]. The active pharmaceutical ingredients of S. miltiorrhiza mainly include lipophilic tanshinones and hydrophilic phenolic acids. The latter is a large group of chemicals, including rosmarinic acid, salvianolic acid A, salvianolic acid B, lithospermic acid, and so forth [2]. Among them, salvianolic acid B is the major component of active phenolic acids in S. miltiorrhiza, while rosmarinic acid, an ester of 3,4-dihydroxyphenyllactic acid and caffeic acid, serves as a basis of salvianolic and lithospermic acids. The biosynthesis of rosmarinic acid requires the coordination of two independent pathways, the phenylpropanoid pathway and the tyrosine-derived pathway. The phenylpropanoid pathway is responsible for the biosynthesis of caffeic acid, whereas the tyrosine-derived pathway is devoted to 3,4-dihydroxyphenyllactic acid biosynthesis (Fig. 1). Key enzymes involved in the two pathways include at least phenylalanine ammonia-lyase (PAL), cinnamic acid 4-hydroxylase (C4H), 4-coumarate:CoA ligase (4CL), tyrosine aminotransferase (TAT), hydroxyphenylpyruvate reductase (HPPR), hydroxyphenylpyruvate dioxygenase (HPPD), and rosmarinic acid synthase (RAS) (Fig. 1).
Phenylalanine ammonia-lyase (EC 4.3.1.5) is the first key enzyme in the phenylpropaniod pathway. It catalyzes the deamination of l-phenylalanine to produce trans-cinnamic acid, which is then converted to p-coumaroyl-CoA by C4H and 4CL for the production of lignin, flavonoid, coumarin, isoflavonoid, furanocoumarin, norlignan, salvianolic acid B, and many other secondary metabolites. PAL was first purified from Hordeum vulgare by Koukol and Connthe [3], but was shown to widely exist in plants, fungi, viruses and algae afterwards. So far, there are no reports for animal PALs. However, it has been shown that PAL can play significant roles in animals and has commercial and medical potential. PAL is able to substantially inhibit neoplastic cell growth in vitro [4] and has the potential to treat the inherited metabolic disorder [5]. Additionally, PAL may be used in large-scale bio-conversion of trans-cinnamic acid and ammonium salts into l-phenylalanine, since the reaction catalyzed by PAL is reversible [6].
Phenylalanine ammonia-lyase is encoded by a small multigene family in most plant species. There are four PAL genes in Arabidopsis thaliana [7, 8], five in Populus trichocarpa [9, 10], three in Scutellaria baicalensis [11], seven in Cucumis sativus [12], and three in Coffea canephora [13]. The members of PAL gene family in a plant are usually expressed differentially in tissues and in response to environmental stimuli and appear to be functionally distinct. For instance, among three C. canephora PALs, CcPAL1 showed high expression in roots, small green-stage beans and pericarps, followed by flowers, red-stage pericarps, branches, large green-stage beans, and less in other tissues analyzed. CcPAL2 showed the highest expression in flowers, followed by pericarps and branches, and less in beans, roots and leaves. Whereas, CcPAL3 was predominantly expressed in small green-stage beans and pericarps, large green- and red-stage pericarps, very low expression in other tissues [13]. Similarly, seven C. sativus PAL genes were also differentially expressed in roots, stems, cotyledons, leaves, flowers and fruits [12]. Analyzing the functions of P. tremuloides PALs showed that PtPAL1 could be involved in the biosynthesis of condensed tannins and other phenolics, whereas PtPAL2 was most likely to be responsible for lignin biosynthesis [14]. However, systematic analysis of the PAL gene family has not been carried out for S. miltiorrhiza.
Recently, a working draft of the S. miltiorrhiza genome has been obtained (Chen et al. unpublished data). In order to get a clear picture of the SmPAL gene family, we searched the current assembly of S. miltiorrhiza genome and then performed PCR amplification of full-length cDNAs. A total of three SmPALs were identified. Gene structures and sequence characteristics of SmPALs were subsequently analyzed. Expression level of SmPAL genes in various tissues of S. miltiorrhiza with or without stress treatment was examined by quantitative real-time RT-PCR. The results suggest the complexity of SmPAL-associated metabolic network and the functional redundancy of SmPAL1 and SmPAL3 in rosmarinic acid biosynthesis. It provides a basis for elucidating the role of SmPAL genes in the biosynthesis of secondary metabolites, particularly the bioactive rosmarinic acid and salvianolic acid B.
Materials and methods
Plant materials and stress treatment
Salvia miltiorrhiza Bunge (line 993), whose genome has been sequenced, was grown in a field nursery. Roots, stems, leaves and flowers were collected from 2-year-old plants in August. Methyl jasmonate (MeJA) treatment was carried out as described previously [15]. Drought treatment was performed by adding 15 % PEG-6000 to 6,7-V liquid media with plantlets pre-cultivated for 2 days. Plantlets were treated for 12, 24, 36 and 48 h and then roots were sampled. Plantlets treated with sterile water were used as controls. The treatments were repeated three times. All samples were frozen immediately after collected and stored in liquid nitrogen until use.
Identification of genomic sequence of SmPAL genes
The genomic sequence of SmPALs were identified by BLAST analysis of four known Arabidopsis AtPALs (NM_129260, NM_115186, NM_120505, NM_111869) against the current assembly of S. miltiorrhiza genome (Chen et al. unpublished data) using the BLASTx algorithm [16]. An e-value cut-off of 10−5 was applied to the homologue recognition. Gene models were predicted as described previously [15].
Cloning of the full-length SmPAL3 cDNA
Total RNA was extracted from roots of S. miltiorrhiza using the general plant total RNA extraction kit (Bioteke, Beijing, China). Poly(A)+ RNA was isolated from total RNA using the Oligotex mRNA purification kit (QIAGEN, MD, CA, USA). Rapid amplification of cDNA ends (RACE) were performed using the GeneRace™ kit (Invitrogen, Carlsbad, CA, USA). PCR amplification of 5′ ends was carried out using the GeneRacer 5′ primer and the nesting gene-specific primer PAL3R1 (5′-CCTGAACTCCTCCACCATCCTCTTCA-3′). Nested PCR amplification of 5′ ends was performed using the GeneRacer 5′ nested primer and the nested gene-specific nested primer PAL3R2 (5′-CTCTGCTTCACGCAGAACCCGTTCT-3′). PCR amplification of 3′ ends was carried out using the GeneRacer 3′ primer and the nesting gene-specific primer PAL3F1 (5′-GAGCAATGGTTTGCTCGTTGATCCT-3′). Nested PCR amplification of 3′ ends was performed using the GeneRacer 3′ nested primer and the nested gene-specific primer PAL3F2 (5′-GGTTTGCTCGTTGATCCTTTGTTGA-3′). PCRs were performed in a kit-recommended 25 μL standard amplification system using 1.5 μL of total first strand cDNA as a template for primary amplification and 0.5 μL of the primary amplification products as a template for nested amplification. The primary amplification reactions for both 5′ and 3′ ends were carried out under the following conditions: predenaturation at 94 °C for 2 min, 5 cycles of amplification at 94 °C for 30 s and 72 °C for 1 min, 5 cycles of amplification at 94 °C for 30 s and 70 °C for 1 min, 25 cycles at 94 °C for 30 s, 60 °C for 45 s and 72 °C for 1.5 min, followed by a final extension at 72 °C for 15 min. The nested amplification reactions for both 5′ and 3′ ends were carried out under the following conditions: predenaturation at 94 °C for 2 min, 30 cycles at 94 °C for 30 s, 60 °C for 45 s and 72 °C for 1.5 min, followed by a final extension at 72 °C for 15 min. PCR products were gel-purified, cloned and then sequenced.
Based on the obtained 5′ and 3′ cDNA sequence, a pair of gene-specific primers, including the forward primer FPAL3 (5′-GTGTGAGCGACTTTCTCTCTCATCT-3′) and the reverse primer RPAL3 (5′-GCGGCTCTCCATTCCACGATTCA-3′), were designed for the amplification of full-length SmPAL3 cDNA. PCR was carried out in a 50 μL volume containing 1 μL cDNA template, 1 μL 10 μM FPAL3, 1 μL 10 μM RPAL3, and 25 μL Premix LA Taq. PCR conditions included 94 °C for 2 min, 35 cycles of 94 °C for 30 s, 58 °C for 45 s and 72 °C for 2.5 min, and 72 °C for 15 min. PCR products were gel-purified, cloned and then sequenced.
Cloning of the 5′ region of SmPAL2 cDNA
5′-RACE was performed using the GeneRace™ kit (Invitrogen) as described above for SmPAL3. cDNA used for primary amplification was transcribed from mRNA isolated from stems of S. miltiorrhiza. The nesting and nested gene-specific primers used in the reactions were PAL2R1 (5′-CCTTGGAGCAATGTGTTGATTCTT-3′) and PAL2R2 (5′-GCAGGGTGTGGTACGAATCACTAT-3′), respectively.
Bioinformatic analysis and phylogenetic tree construction
The molecular weight (MW) and theoretical isoelectric point (pI) were predicted using the Compute pI/MW tool on the ExPASy server [17] (http://web.expasy.org/compute_pi/). Known PAL sequences shown high identities with the deduced amino acid sequence of SmPALs were identified by BLASTP analyses of SmPALs against the database of non-redundant protein sequences in NCBI using the default alignment parameters (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The alignment of deduced amino acid sequences of SmPALs and PALs from other organisms was performed using ClustalW version 1.83 using the default parameters [18]. Phylogenetic tree was constructed using MEGA version 4.0 [19]. The reliability of branching was assessed by the bootstrap re-sampling method using 1,000 bootstrap replicates and only nodes supported by bootstrap values >50 % are shown.
Quantitative real-time PCR
Total RNA was isolated from the tissues of S. miltiorrhiza with or without stress treatment using the general plant total RNA extraction kit (Bioteke, Beijing, China) and then digested with RNase-free DNase I to remove the genomic DNA contamination. Reverse transcription was carried out by SuperScript III Reverse Transcriptase. The reaction was achieved by incubating at 65 °C for 5 min, 50 °C for 1 h, and 70 °C for 15 min as described [20]. The resulting cDNA was diluted to 500 μL with sterile water. Quantitative Real-time PCR was carried out in triplicate reactions using the BIO-RAD CFX system. Gene-specific primers used were GATCTCTTCACGGAAGACCGTTGAA and GCCATTGACGCCCATTGTGAGAGTT for SmPAL1, CAGGATCAAGGGGAGCAGATCGTA and GCACCATTCCATTCCCTGAGACA for SmPAL2, and CCCGCGATCGGGAACAGGATCAA and GCGGCTCTCCATTCCACGATTCA for SmPAL3. Ubiquitin gene was used as a reference as described previously [15]. The forward and reverse primers for amplification of ubiquitin were AGATGGGCGGACACTTGCTGATTA and ACTCTCCACCTCCAAAGTGATGGT, respectively. PCRs were carried out in a 20 μL volume containing 2 μL diluted cDNA, 10 μM forward primer, 10 μM reverse primer and 1 × SYBR Premix Ex TaqII using the following conditions: 95 °C for 30 s, 39 cycles of 95 °C for 5 s, 60 °C for 18 s and 72 °C for 15 s. Relative abundance of transcripts was determined using the comparative Cq method [21]. For analyzing the responses of SmPALs to MeJA and PEG-6000 treatments, the expression of SmPALs in roots without treatment was used as controls and was arbitrarily set to 1. Standard deviations were calculated from three PCR replicates.
Results
Genome-wide identification and molecular cloning of SmPALs
The decoding of the S. miltiorrhiza genome (Chen et al. unpublished data) enables us to perform a genome-wide search of SmPALs through BLAST analysis of four known Arabidopsis AtPALs (NM_129260, NM_115186, NM_120505, NM_111869) against the current assembly of S. miltiorrhiza genome using the BLASTx algorithm [16]. As a result, we identified a total of three genomic loci of PAL genes. It includes the known SmPAL1 (accession nos. EF462460 for genomic DNA and DQ408636 for cDNA) [22–24], the genomic sequence of SmPAL2 with only partial cDNA sequence available (accession no. GQ249111) and the third PAL gene, SmPAL3, which has never been reported. We then cloned the full-length SmPAL3 cDNA and the 5′ region of SmPAL2 cDNA using PCR technology. Taken together, it suggests that both the genomic sequence and the cDNA sequence of three SmPALs have been obtained and are available for comparative analysis.
Analysis of cDNA sequence of SmPALs showed that the open reading frames (ORFs) of SmPAL1, SmPAL2 and SmPAL3 are 2,133, 2,127, and 2,127 bp, respectively, suggesting SmPALs are highly similar in the length of ORF regions (Table 1). However, substantial difference was observed for sequence identity among them. SmPAL1 and SmPAL3 shares 85 % sequence identity, whereas the identity between SmPAL1 and SmPAL2 and between SmPAL2 and SmPAL3 is only 75 and 77 %, respectively, suggesting SmPAL2 has less homology to SmPAL1 and SmPAL3 than that between SmPAL1 and SmPAL3. Comparison analysis of genomic sequence and cDNA sequence showed that all of three SmPAL genes contain only one intron and the exon-intron junctions satisfy the GT-AG rule for donor/acceptor sites (Fig. 2). The intron is located at 386, 380 and 380 bp from ATG of SmPAL1, SmPAL2, and SmPAL3, respectively. The first exon of SmPAL1 includes 387 bp coding sequence, whereas the coding sequence in the first exon of both SmPAL2 and SmPAL3 is 381 bp in length. The length of coding sequence in the second exon (1,750 bp) is conserved among three SmPAL genes. The predicted MW and theoretical pI of SmPAL1, SmPAL2 and SmPAL3 proteins are 77.1 kDa and 6.06, 77.2 kDa and 5.98, 76.5 kDa and 5.96, respectively (Table 1), which are consistent with previous results showing the MW of plant PALs varied from 72 to 83 kDa [25].
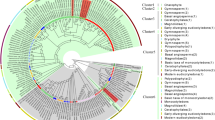
The deduced amino acid sequence of SmPAL1, SmPAL2 and SmPAL3 includes 711, 709, and 709 amino acids, respectively, and shares 80 % identity between SmPAL1 and SmPAL2, 79 % between SmPAL2 and SmPAL3, and 90 % between SmPAL1 and SmPAL3 (Fig. 3). It suggests that SmPAL1 and SmPAL3 are highly homologous, whereas SmPAL2 has relatively low homology with both SmPAL1 and SmPAL3, which is consistent with the results from cDNA sequence analysis of SmPALs. Additionally, SmPALs share over 80 % identity with many PALs from other plant species, such as Melissa officinalis MoPAL [26], Agastache rugosa ArPAL [27], Perilla frutescens PfPAL, S. baicalensis SbPAL1-3 [11], S. viscidula SvPAL [28], and A. thaliana AtPAL1-4 [7, 8], confirming the role of the identified SmPALs in encoding PAL in S. miltiorrhiza. In order to further characterize SmPALs, we constructed a phylogenetic tree of three SmPALs and 57 PALs from other organisms. As shown in Fig. 4, PALs from fungus, gymnosperms, monocots and dicots clustered to four clades and PALs from dicots might be divided into four sub-clades. SmPAL1, SmPAL2 and SmPAL3 were most closely related to ArPAL, SbPAL1, and SvPAL, respectively. Like S. miltiorrhiza, A. rugosa and S. baicalensis and S. viscidula are members of the Lamiaceae family and are perennial herbs. SmPAL1 and SmPAL3 clustered to a sub-clade with rosmarinic acid biosynthesis-related MoPAL [26], Cistanche deserticola CdPAL associated with the biosynthesis of phenolic compounds [29], Rehmannia glutinosa RgPAL responsive to oxidative stress [30], and various PALs with unknown functions, whereas SmPAL2 clustered to the other sub-clade with several function-unknown PALs (Fig. 4). It suggests the sequence diversity of SmPAL2 from SmPAL1 and SmPAL3.
Joined phylogenetic tree of PALs from S. miltiorrhiza and other organisms. The deduced full-length amino acid sequences were aligned using ClustalW version 1.83 and the phylogenetic tree was constructed using MEGA 4.0 by the neighbor-joining (NJ) method with 100 bootstrap replicates. PALs from fungus, gymnosperms, monocots and dicots are indicated with backgrounds in purple, blue, red, and green, respectively. SmPALs are highlighted with yellow. Ar Agastache rugosa, At Arabidopsis thaliana, Bo Bambusa oldhamii, Ca Coffea arabica, Cc Coffea canephora, Cd Cistanche deserticola, Cl Citrus limon, Cr Catharanthus roseus, Cs Camellia sinensis, Dc Daucus carota, Dl Digitalis lanata, Ep Euphorbia pulcherrima, Gb Ginkgo biloba, In Ipomoea nil, Jc Jatropha curcas, Me Manihot esculenta, Mo Melissa officinalis, Na Nicotiana attenuata, Nt Nicotiana tabacum, Os Oryza sativa, Pf Perilla frutescens, Pp Pinus pinaster, Pt Populus trichocarpa, Pta Pinus taeda, Rg Rehmannia glutinosa, Rt Rhodosporidium toruloides, Sb Scutellaria baicalensis, Sv Scutellaria viscidula, Ta Triticum aestivum, Vv Vitis vinifera, Zm Zea mays. (Color figure online)
Comparative analysis of the 5′-flanking regions of SmPAL genes
In order to investigate the transcriptional activity of three SmPAL gene promoters, 750 bp 5′-flanking sequence of the coding region was extracted from each SmPAL gene and analyzed using the PLACE (http://www.dna.Affrc.go.jp/PLACE/) and PlantCARE databases (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to identify putative cis-elements [31, 32]. The results showed that there were several cis-elements conserved in the promoters of three SmPALs. It includes the box L (YCYYACCWACC), AC element (CYCACCWACC), MYB2 recognition site (YAACKG), E-box (CANNTG), and so on (Table 2). Box L is one of the three motifs (box P, YTYYMMCMAMCMMC; box A, CCGTCC; and box L) previously shown to exist in plant PAL, C4H and 4CL gene promoters [33]. Box L is tightly co-regulated with box A and box P and appear to be necessary but not sufficient for elicitor- or light-mediated gene activation in parsley [33]. However, in S. miltiorrhiza, box A exist only in the promoter of SmPAL3 and no box P was found in all SmPAL gene promoters (Table 2). The absence of one or two of box L and box A and box P was also observed in the promoters of P. trichocarpa and P. tremuloides C4H genes with distinct physiological functions [34]. It indicates that the different combination of these boxes may result in a remarkable degree of variability in gene functions. The sequences of AC element and box L are overlapped and appear to play similar roles. MYB2 recognition site (YAACKG) is a cis-acting element associated with drought-induced gene expression [35]. The presence of this element in all of the SmPAL gene promoters indicates SmPALs to be drought-induced. E-box is a cis-element recently found to be responsible for inducing gene expression in response to MeJA treatment [36]. Examination of the promoters of SmPALs revealed that there are four E-box cis-elements in SmPAL2 promoter, whereas the number of E-box cis-element in SmPAL1 and SmPAL3 was only one (Table 2). It suggests that all three SmPALs are probably induced by MeJA treatment to various degrees.
In addition to the elements conserved among three SmPALs, many elements were found only in the promoters of one or two SmPALs. For instance, AG motif (AGATCCAA), a cis-element sufficient to confer wounding and elicitor responsiveness [37], exists only in the promoter of SmPAL1 (Table 2). These results indicate that SmPALs may commonly response to some biotic and abiotic stresses, whereas in the other cases, the response can be differential for individual SmPAL. The underlying mechanisms of SmPALs in response to stress are complicated and need to be further investigated.
Tissue-specific expression of SmPAL genes
The expression levels of SmPAL1, SmPAL2 and SmPAL3 in various S. miltiorrhiza tissues, including roots, stems, leaves and flowers, were analyzed using qRT-PCR. The transcripts of all three SmPAL genes could be detected in all of the tissues analyzed, but showed differential expression levels (Fig. 5). SmPAL1, the most abundant among three SmPALs in all of the tissues analyzed, showed the highest expression in roots, followed by leaves, stems and flowers. SmPAL2 was predominately expressed in stems and flowers. Its expression levels in roots and leaves were very low. SmPAL3 had the highest expression in leaves, less in roots, stems and flowers. The results suggest SmPAL1 and SmPAL3 can function in all of the tissues analyzed, whereas SmPAL2 appears to play roles mainly in stems and flowers under normal conditions.
Responses of SmPAL genes to PEG-6000 and MeJA treatments
The presence of many environmental signal-responsive cis-elements in the promoters of SmPAL genes suggests that SmPAL genes might be involved in plant response to stress. To test this hypothesis, we analyzed the expression level of SmPAL genes in roots of plantlets treated with PEG-6000 and MeJA using qRT-PCR. The results showed that all three SmPALs were regulated by drought and MeJA treatments, although the time and degree of reaction differed from one another (Fig. 6). Under drought conditions, SmPAL1 and SmPAL3 were up-regulated to about threefolds after being treated for 24 and 36 h, respectively (Fig. 6). The up-regulated level of SmPAL2 after 36 h drought treatment was the most significant, which reached to about ninefolds (Fig. 6). Similar results were also observed for SmPALs in roots of plantlets treated with MeJA for 24 and 36 h (Fig. 6). However, down-regulation was found for all SmPALs after 12 h MeJA treatment and SmPAL1 was up-regulated against after 48 h treatment in addition to the up-regulation after 24 h treatment, which were different from the results observed for drought treatment (Fig. 6). These results suggest that all of three SmPALs are drought- and MeJA- responsive. Among them, SmPAL1, which was induced significantly in plantlets treated for 24 h, showed the quickest response to both drought and MeJA treatments, whereas SmPAL2, which was induced to about nine and 18-folds after 36 h treatment, had the most significant response to these treatments. Considering the low expression of SmPAL2 in roots of plants cultivated under normal conditions, the significant change of SmPAL2 transcripts could be important for plant response to environmental stress.
Discussion
PAL is a significant key enzyme in the biosynthesis of many useful secondary metabolites. In this study, we successfully performed a genome-wide search of SmPAL genes by BLAST analysis of the current assembly of S. miltiorrhiza genome (Chen et al. unpublished data). A total of three SmPAL genes were identified. Using PCR technology, we cloned the full-length SmPAL3 cDNA and the 5′-region of SmPAL2 cDNA that were unknown previously. The capability of the identified genes in encoding PAL in S. miltiorrhiza was confirmed by high sequence similarity between SmPALs and known PALs from A. thaliana [8], M. officinalis [26], A. rugosa [27], P. frutescens, S. baicalensis [11], S. viscidula [28], and so on. The existence of a small multigene family in S. miltiorrhiza is consistent with the results from other plant species with whole genome sequence available [8–10, 12].
Phylogenetic analysis of SmPALs and PALs from other organisms showed that PALs from fungus, gymnosperms, monocots and dicots clustered to four distinct clades and PALs from dicots might be divided into four sub-clades (Fig. 4). The identified three SmPALs fell into two different sub-clades of dicot PALs with SmPAL1 and SmPAL3 in a sub-clade whereas SmPAL2 in the other one. Consistently, the identity of SmPAL1 and SmPAL3 at both the nucleotide sequence level and the amino acid sequence level is higher than that of SmPAL1 and SmPAL2 and of SmPAL2 and SmPAL3. It indicates the role of SmPAL2 is different from SmPAL1 and SmPAL3 in some aspects in S. miltiorrhiza. Since SmPAL3 clustered in a sub-clade with MoPAL and SmPAL1 that were involved in the biosynthesis of rosmarinic acid [24, 26] and CdPAL that was associated with the biosynthesis of phenolic compounds [29], it is very likely that SmPAL3 is also involved in rosmarinic acid biosynthesis in S. miltiorrhiza. However, it is currently unknown whether SmPAL2 is involved in rosmarinic acid biosynthesis because there is no information about the role of PALs clustered with SmPAL2 in the biosynthesis of rosmarinic acid.
It has been shown that different member of the PAL gene family in a plant may have distinguishable biochemical, molecular and catalytic properties [38] and involved in the production of different products under specific conditions [39]. In coffee bean, CcPAL1 and CcPAL3 are associated with the accumulation of cholorogenic acids (CGA), whereas CcPAL2 may contribute more significantly to flavonoid accumulation [13]. Similar results were also observed for two P. tremuloides PALs, of which PtPAL1 was involved in condensed tannin metabolism and PtPAL2 was associated with monolignol biosynthesis [14]. Consistently, differential expression patterns were observed for three SmPALs, although they were expressed in all of the tissues analyzed (Fig. 5). The transcripts of SmPAL1 and SmPAL3 showed high levels in roots and leaves, whereas SmPAL2 were predominately expressed in stems and flowers. It indicates that specific products of the phenylpropaniod pathway are probably synthesized through a metabolic channel organized by specific isoenzymes. However, it has also been shown that different PAL genes in a plant may coordinately function together in the production of a specific product. For instance, A. thaliana AtPAL1, AtPAL2 and AtPAL4 were associated with lignin biosynthesis, and AtPAL1 and AtPAL2 were also involved in the biosynthesis of stress-induced flavonoids [8, 40, 41]. In S. baicalensis, all of three SbPAL genes were associated with stress-induced flavonoid synthesis [11]. In this study, SmPAL1 and SmPAL3 were found to cluster with SbPAL2 and SbPAL3 in a sub-clade of dicot PALs, and SmPAL2 clustered with SbPAL1 in the other sub-clade (Fig. 4). It indicates that all three SmPALs may function redundantly as SbPALs in the biosynthesis of flavonoids in S. miltiorrhiza.
Analysis of the 5′-flanking regions of SmPAL genes showed the existence of several conserved cis-elements, such as elicitor- or light-associated box L and AC element (CYCACCWACC) [33], drought-related MYB2 recognition site [35] and MeJA treatment-responsive E-box [36], suggesting that SmPALs could be functionally redundant in response to some environment stresses in S. miltiorrhiza (Table 2). Consistently, the expression of SmPALs was all induced in roots of plantlets treated with PEG-6000 and MeJA (Fig. 6). On the other hand, difference was observed for three SmPALs in the time and degree of response after PEG-6000 and MeJA treatments. Similar results were previously found for three PALs from S. baicalensis [11]. All three SbPALs responded to MeJA treatment, but their responses were different. SbPAL1 showed the highest induced by 100 μM MeJA, whereas SbPAL2 and SbPAL3 were induced to the highest degree by 200 μM MeJA. In the tissues treated with 100 μM MeJA, SbPAL1 and SbPAL2 showed the highest induction in 48 h, while SbPAL3 reached to the highest point after 12 h treatment [11]. Because many of the downstream products of the phenylpropaniod pathway, such as lignins, flavonoids, coumarins, isoflavonoids, furanocoumarins and norlignans, are cell wall constituents, pigments, UV protectants and plant defense compounds [42–44], the subtle regulation of PAL gene expression could be very important for plants to adapt to the stressful environments.
Taken together, our results showed the existence of at least three SmPALs in S. miltiorrhiza. These SmPALs may function redundantly in the biosynthesis of some metabolites, but the roles of each SmPAL maybe different in the biosynthesis of other metabolites, suggesting the complexity of SmPAL-associated metabolic network in S. miltiorrhiza. Further analyzing the physiological functions of SmPALs using transgenic approaches will definitely shed light on elucidating the metabolic network of secondary metabolites in S. miltiorrhiza.
Abbreviations
- 4CL:
-
4-Coumarate:CoA ligase
- C4H:
-
Cinnamic acid 4-hydroxylase
- HPPD:
-
Hydroxyphenylpyruvate dioxygenase
- HPPR:
-
Hydroxyphenylpyruvate reductase
- MeJA:
-
Methyl jasmonate
- MW:
-
Molecular weight
- ORF:
-
Open reading frame
- PAL:
-
Phenylalanine ammonia-lyase
- PCR:
-
Polymerase chain reactions
- pI :
-
Isoelectric point
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- RACE:
-
Rapid amplification of cDNA ends
- RAS:
-
Rosmarinic acid synthase
- TAT:
-
Tyrosine aminotransferase
- TCM:
-
Traditional Chinese medicine
References
Cheng TO (2006) Danshen: a popular Chinese cardiac herbal drug. J Am Coll Cardiol 47:1498
Wang XH, Morris-Natschke SL, Lee KH (2007) Developments in the chemistry and biology of the bioactive constituents of Tanshen. Med Res Rev 27:133–148
Koukol J, Conn EE (1961) The metabolism of aromatic compounds in higher plants. IV. Purification and properties of the phenylalanine deaminase of Hordeum vulgare. J Biol Chem 236:2692–2698
Abell CW, Stith WJ, Hodgins DS (1972) The effects of phenylalanine ammonia-lyase on leukemic lymphocytes in vitro. Cancer Res 32:285–290
Hyun MW, Yun YH, Kim JY, Kim SH (2011) Fungal and plant phenylalanine ammonia-lyase. Mycobiology 39:257–265
Hamilton BK, Hsiao HY, Swann WE, Anderson DM, Delent JJ (1985) Manufacture of l-amino acids with bioreactors. Trends Biotechnol 3:64–68
Wanner LA, Li G, Ware D, Somssich IE, Davis KR (1995) The phenylalanine ammonia-lyase gene family in Arabidopsis thaliana. Plant Mol Biol 27:327–338
Raes J, Rohde A, Christensen JH, Van de Peer Y, Boerjan W (2003) Genome-wide characterization of the lignification toolbox in Arabidopsis. Plant Physiol 133:1051–1071
Shi R, Sun YH, Li Q, Heber S, Sederoff R, Chiang VL (2010) Towards a systems approach for lignin biosynthesis in Populus trichocarpa: transcript abundance and promoter sequence motifs of the monolignol biosynthetic genes. Plant Cell Physiol 51:144–163
Shi R, Yang C, Lu S, Sederoff R, Chiang VL (2010) Specific downregulation of PAL genes by artificial microRNAs in Populus trichocarpa. Planta 232:1281–1288
Xu H, Park NI, Li X, Kim YK, Lee SY, Park SU (2010) Molecular cloning and characterization of phenylalanine ammonia-lyase, cinnamate 4-hydroxylase and genes involved in flavone biosynthesis in Scutellaria baicalensis. Bioresour Technol 101:9715–9722
Shang QM, Li L, Dong CJ (2012) Multiple tandem duplication of the phenylalanine ammonia-lyase genes in Cucumis sativus. Planta 236:1093–1105
Lepelley M, Mahesh V, McCarthy J, Rigoreau M, Crouzillat D, Chabrillange N, de Kochko A, Campa C (2012) Characterization, high-resolution mapping and differential expression of three homologous PAL genes in Coffea canephora Pierre (Rubiaceae). Planta 236:313–326
Kao YY, Harding SA, Tsai CJ (2002) Differential expression of two distinct phenylalanine ammonia-lyase genes in condensed tannin-accumulating and lignifying cells of quaking aspen. Plant Physiol 130:796–807
Ma Y, Yuan L, Wu B, Li X, Chen S, Lu S (2012) Genome-wide identification and characterization of novel genes involved in terpenoid biosynthesis in Salvia miltiorrhiza. J Exp Bot 63:2809–2823
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Bjellqvist B, Basse B, Olsen E, Celis JE (1994) Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis 15:529–539
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Wu B, Li Y, Yan H, Ma Y, Luo H, Yuan L, Chen S, Lu S (2012) Comprehensive transcriptome analysis reveals novel genes involved in cardiac glycoside biosynthesis and mlncRNAs associated with secondary metabolism and stress response in Digitalis purpurea. BMC Genomics 13:15
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25(4):402–408
Hu YS, Zhang L, Di P, Chen WS (2009) Cloning and induction of phenylalanine ammonia-lyase gene from Salvia miltiorrhiza and its effect on hydrophilic phenolic acids levels. Chin J Nat Med 7:0449–0457
Song J, Wang Z (2009) Molecular cloning, expression and characterization of a phenylalanine ammonia-lyase gene (SmPAL1) from Salvia miltiorrhiza. Mol Biol Rep 36:939–952
Song J, Wang Z (2011) RNAi-mediated suppression of the phenylalanine ammonia-lyase gene in Salvia miltiorrhiza causes abnormal phenotypes and a reduction in rosmarinic acid biosynthesis. J Plant Res 124:183–193
Hahlbrock K, Grisebach H (1979) Enzymatic controls in biosynthesis of lignin and flavonoids. Annu Rev Plant Physiol 30:105–130
Weitzel C, Petersen M (2010) Enzymes of phenylpropanoid metabolism in the important medicinal plant Melissa officinalis L. Planta 232:731–742
Tuan PA, Park WT, Xu H, Park NI, Park SU (2012) Accumulation of tilianin and rosmarinic acid and expression of phenylpropanoid biosynthetic genes in Agastache rugosa. J Agric Food Chem 60:5945–5951
Lei W, Yao RX, Kang XH, Tang SH, Qiao AM, Sun M (2011) Isolation and characterization of the anthocyanidin genes PAL, F3H and DFR of Scutellaria viscidula (Lamiaceae). Genet Mol Res 10:3385–3402
Hu GS, Jia JM, Hur YJ, Chung YS, Lee JH, Yun DJ, Chung WS, Yi GH, Kim TH, Kim DH (2011) Molecular characterization of phenylalanine ammonia lyase gene from Cistanche deserticola. Mol Biol Rep 38:3741–3750
Lee BK, Park MR, Srinivas B, Chun JC, Kwon IS, Chung IM, Yoo NH, Choi KG, Yun SJ (2003) Induction of phenylalanine ammonia-lyase gene expression by paraquat and stress-related hormones in Rehmannia glutinosa. Mol Cells 16:34–39
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res 27:297–300
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Logemann E, Parniske M, Hahlbrock K (1995) Modes of expression and common structural features of the complete phenylalanine ammonia-lyase gene family in parsley. Proc Natl Acad Sci USA 92:5905–5909
Lu S, Zhou Y, Li L, Chiang VL (2006) Distinct roles of cinnamate 4-hydroxylase genes in Populus. Plant Cell Physiol 47:905–914
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78
Miyamoto K, Shimizu T, Lin F, Sainsbury F, Thuenemann E, Lomonossoff G, Nojiri H, Yamane H, Okada K (2012) Identification of an E-box motif responsible for the expression of jasmonic acid-induced chitinase gene OsChia4a in rice. J Plant Physiol 169:621–627
Sugimoto K, Takeda S, Hirochika H (2003) Transcriptional activation mediated by binding of a plant GATA-type zinc finger protein AGP1 to the AG-motif(AGATCCAA) of the wound-inducible Myb gene NtMyb2. Plant J 36:550–564
Kumar A, Ellis BE (2001) The phenylalanine ammonia-lyase gene family in raspberry. Structure, expression, and evolution. Plant Physiol 127:230–239
Camm EL, Towers GHN (1973) Phenylalanine ammonia lyase. Phytochemistry 1973(12):961–973
Rohde A, Morreel K, Ralph J, Goeminne G, Hostyn V, De Rycke R, Kushnir S, Van Doorsselaere J, Joseleau JP, Vuylsteke M, Van Driessche G, Van Beeumen J, Messens E, Boerjan W (2004) Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on and carbohydrate metabolism. Plant Cell 16:2749–2771
Olsen KM, Lea US, Slimestad R, Verheul M, Lillo C (2008) Differential expression of four Arabidopsis PAL genes; PAL1 and PAL2 have functional specialization in abiotic environmental-triggered flavonoid synthesis. J Plant Physiol 165:1491–1499
Whitbred JM, Schuler MA (2000) Molecular characterization of CYP73A9 and CYP82A1 P450 genes involved in plant defense in pea. Plant Physiol 124:47–58
Suzuki S, Nakatsubo T, Umezawa T, Shimada M (2002) First in vitro norlignan formation with Asparagus officinalis enzyme preparation. Chem Commun 2002:1088–1089
Suzuki S, Yamamura M, Shimada M, Umezawa T (2004) A heartwood norlignan, (E)-hinokiresinol, is formed from 4-coumaryl 4-coumarate by a Cryptomeria japonica enzyme preparation. Chem Commun 2004:2838–2839
Acknowledgments
We thank Dr. Shilin Chen and the sequencing group in our institute for kindly providing the S. miltiorrhiza genome sequence. This work was supported by grants from the Beijing Natural Science Foundation (Grant No. 5112026 to SL), the Major Scientific and Technological Special Project for Significant New Drugs Creation (Grant No. 2012ZX09301002-001-031 to SL), the Program for Changjiang Scholars and Innovative Research Team in University (IRT1150 to SL), the Research Fund for the Doctoral Program of Higher Education of China (20111106110033 to SL), and the Program for Xiehe Scholars in Chinese Academy of Medical Sciences & Peking Union Medical College (to SL).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hou, X., Shao, F., Ma, Y. et al. The phenylalanine ammonia-lyase gene family in Salvia miltiorrhiza: genome-wide characterization, molecular cloning and expression analysis. Mol Biol Rep 40, 4301–4310 (2013). https://doi.org/10.1007/s11033-013-2517-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-013-2517-3