Abstract
Genotype-phenotype correlation of hypertrophic cardiomyopathy (HCM) has been challenging because of the genetic and clinical heterogeneity. To determine the mutation profile of Chinese patients with HCM and to correlate genotypes with phenotypes, we performed a systematic mutation screening of the eight most commonly mutated genes encoding sarcomere proteins in 200 unrelated Chinese adult patients using direct DNA sequencing. A total of 98 mutations were identified in 102 mutation carriers. The frequency of mutations in MYH7, MYBPC3, TNNT2 and TNNI3 was 26.0, 18.0, 4.0 and 3.5 % respectively. Among the 200 genotyped HCM patients, 83 harbored a single mutation, and 19 (9.5 %) harbored multiple mutations. The number of mutations was positively correlated with the maximum wall thickness. We found that neither particular gene nor specific mutation was correlated to clinical phenotype. In summary, the frequency of multiple mutations was greater in Chinese HCM patients than in the Caucasian population. Multiple mutations in sarcomere protein may be a risk factor for left ventricular wall thickness.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hypertrophic cardiomyopathy (HCM) is a disease marked by left ventricular hypertrophy with predominant involvement of the interventricular septum in the absence of other causes of hypertrophy. HCM is the most prevalent, heritable cardiovascular disease and the most common cause of sudden cardiac death in young athletes [1]. We have previously reported that the prevalence of HCM is 83/100,000 in the adult Chinese population, indicating that there are approximately one million HCM patients in China [2].
Over 1400 mutations in at least 11 genes encoding sarcomere proteins have been reported to cause HCM [3, 4]. Currently, 3–5 % of patients with HCM have been estimated to carry more than one mutation in the same gene or different genes. These patients are thought to have severer clinical manifestations than did one mutation carrier. These studies were mostly performed in Caucasians and little is known about the genetic basis on multiple mutations in Chinese patients with HCM. To assess the mutation profile and the genotype-phenotype correlations, systematic mutation screening of the eight most common HCM-disease genes encoding the sarcomere proteins was carried out in 200 unrelated index Chinese patients with HCM. These HCM-causing genes encode beta-myosin heavy chain (MYH7), cardiac myosin binding protein C (MYBPC3), the regulatory and essential myosin light chains (MYL2, MYL3), alpha-tropomyosin (TPM1), cardiac troponin I (TNNI3), cardiac troponin T (TNNT2) and alpha-actin (ACTC1).
Materials and methods
Subjects
Two hundred unrelated index patients with HCM and 120 age-and sex-matched healthy controls were recruited consecutively from Beijing Fuwai Hospital, Chinese Academy of Medical Sciences, from 2002 to 2008. This study was carried out in accordance with the Declaration of Helsinki (2000) of the World Medical Association [5], and has been approved by the Ethics Committee of Fuwai Hospital. Written informed consent was provided by all participants. The diagnosis of HCM was ascertained in adults by a left ventricular maximal wall thickness (MWT) of greater than 15 mm on echocardiography [6]. The greatest wall thickness measured in diastole at any site in the LV wall was regarded as the maximal thickness, independent of correction for body surface area, gender or age. Subjects with extracardiac disease or secondary causes of cardiac hypertrophy were excluded. None of the control subjects had a history of serious systemic diseases. Three cardiologists independently reviewed all clinical data.
Mutation detection
Genomic DNA was prepared from peripheral blood leukocytes. Mutation screening was performed on the entire coding sequences and flanking regions in MYH7, MYBPC3, MYL2, MYL3, TPM1, TNNI3, TNNT2 and ACTC1 with polymerase chain reaction (PCR) amplification and direct DNA sequencing on an ABI Prism 3730 XL DNA Sequencer (Applied Biosystems, Foster City, CA, USA). All variants were confirmed by sequencing in sense and anti-sense directions. Non-pathogenic polymorphisms, defined as more than 1 % frequency in 120 unrelated control subjects, were excluded. The pathological variants were defined as mutation-HCM cosegregation in all affected members in the investigated pedigrees and the gene has been reported as a HCM-causing gene in literature; the mutation is localized in a very conservative sequence region across species; and the mutation causes a significant structural or functional change by bioinformatic prediction [7, 8]. To confirm the pathogenic role for a mutation, the following programs were applied. The programs of amino acid substitution prediction (SIFT, http://sift.bii.a-star.edu.sg/www/SIFT_BLink_submit.html; Polyphen-HCM, http://genetics.bwh.harvard.edu/pph/ or Polyphen-2 when prediction with Polyphen-HCM unavailable, http://genetics.bwh.harvard.edu/pph2/; Mutpred, http://mutpred.mutdb.org/; SNPs3D, http://www.snps3d.org/; Pmut, http://mmb2.pcb.ub.es:8080/PMut/). A missense mutation was assumed to be possibly disease-causing if at least two independent programs indicated a damaging effect. The program Human Splicing Finder (HSF) was used to determine whether any of the detected mutations destroys an existing or creates a novel splice site (HSF, http://www.umd.be/HSF/).
Statistical analysis
Statistical analysis was performed with SPSS for Windows, release 18.0 (SPSS, Chicago, IL). Continuous variables were expressed as mean ±S.D. ANOVA analysis was used to calculate the difference between groups. Qualitative parameters were compared between groups by Chi squared test or Fisher’s exact test. A two-tailed P value of <0.05 was considered significant. To rule out the influence of gender or age, the General Linear Model was introduced to adjust the differences in clinical characteristics among the groups.
Results
Genotype analysis
Among the 200 patients with HCM, 102 (51 %) were identified with the disease-causing mutations in the genotyped 8 genes. Mutations were detected in 61 % (58/95) of the familial probands and 41.9 % (44/105) of the sporadic cases. Seventy (47.9 %) male and 32 (59.3 %) female patients were found to harbor mutations, respectively.
A total of 98 mutations were identified in 102 mutation carriers (Table 1), 58 % (57/98) of these mutations were novel mutation. The novel mutation was defined as not reported in the previous publications or the mutation databases (http://www.cardiogenomics.org and http://www.hgmd.org). The mutation was distributed mostly in MYH7 (26.0 %, 52/200) and in MYBPC3 (18.0 %, 36/200), less in TNNT2 (4.0 %, 8/200) and in TNNI3 (3.5 %, 7/200). Mutations no more than 1.5 % was found in each of the following genes, including MYL2 (1 %, 2/200), MYL3 (1.5 %, 3/200), TPM1 (1.5 %, 3/200) and ACTC1 (1.5 %, 3/200), as shown in the Fig. 1. Various types of mutations were detected in MYBPC3, including missense, splicesite, nonsense and frame shift mutation. In contrast, missense mutations were predominantly identified in the genes other than MYBPC3.
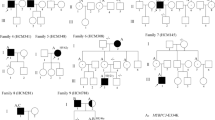
Among the 102 HCM mutation carriers, 83 had a single mutation identified, 18 (9 %) had two mutations, and 1 had three mutations, respectively (Table 2). Eight of the double mutations were located within a single gene (MYH7 or MYBPC3), whereas the other double mutations and the triple mutations presented in distinct genes.
HCM phenotype
Nineteen patients were found to carry multiple mutations. The effect of the number of mutations on the phenotype was further analyzed. Patients were classified into three groups on the basis of mutation number: patients with no mutation (98 patients), single mutation (83 patients), and multiple mutations (≥2 mutations) (19 patients). MWT was proportionally related to the mutation numbers (Table 3), even after adjusting for age and gender. MWT was increased with the numbers of mutations harbored (non-mutation vs. 1 vs. 2 or more, 19.7 ± 5.1 mm vs. 20.5 ± 4.8 mm vs. 23.6 ± 5.7 mm, P < 0.01) and earlier age at diagnosis (non-mutation vs. 1 vs. 2 or more, 44.9 ± 13.5 years vs. 38.4 ± 12.7 years vs. 34.5 ± 12.9 years, P < 0.01). The left atrial internal diameter (LA) was greater in patients with mutations than in those without mutations (LA, non-mutation vs. 1 vs. 2 or more, 38.4 ± 6.3 mm vs. 41.4 ± 8.3 mm vs. 42.5 ± 7.9 mm, P < 0.01). No significant differences in left ventricular end-diastolic diameter (LVEDD) or left ventricular ejection fraction (LVEF) were observed among the three groups. Of the 19 patients with multiple mutations, 11 carried at least one established risk markers of SCD and 1 was treated with implantable cardioverter defibrillator (ICD).
Discussion
We report the mutation profiling of eight genes encoding sarcomere proteins in an adult Chinese HCM cohort. As most reports, the gene mutations can be found in more than half of the HCM patients. The MYH7 (26.0 %) and MYBPC3 (18.0 %) were the predominant HCM-causing genes in Chinese population as well, followed by TNNT2 (4.0 %) and TNNI3 (3.5 %). The other four screened genes accounted for less than 1.5 % each. In addition to missense mutation, splice site, nonsense and frame shift mutations could also be identified in MYBPC3, but not in the other screened genes.
The gene mutations, such as R403Q in MYH7, have been found as malignant mutation in Caucasian HCM [9, 10], but were not detected in Chinese. To date, more than 1,400 mutations in at least 11 HCM-causing sarcomere genes have been identified in patients with HCM. Genotype-phenotype correlations have been challenging because of the genetic and clinical heterogeneity of HCM, such as that most pedigrees carried their own private mutation. Multiple mutations were found in Caucasians, the frequency of multiple mutations of two or more in the same gene or in different genes has been reported to be only 3–6 and 0.8 %, respectively [11–16]. Only 2.7 % (3/112) familial HCM patients were found harbor compound gene mutations in the selected eight genes in Japanese [17]. In contrast, we screened 8 known HCM-causing genes in 200 HCM patients, 83 had a single mutation, 18 (9 %) had two mutations, and 1 had three mutations. The frequency of multiple mutations was much higher in this study than in previous reports [11–16].
The frequency of mutation in adult proband (61.0 %, 58/95) and sporadic patients (41.9 %, 44/105) were similar to that of previously reported in HCM children (64 % in familial and 49 % in sporadic, respectively) [18]. Both familial and sporadic patients of HCM shared common gene mutations, indicating identical feasibility and necessity of genetic analyses for both types of patients, and in both children and adults.
No genetic mutation was identified in 49.0 % (98/200) of HCM patients. Three conceivable reasons maybe responsible: mutations were present in genes that were not screened; mutations were present in the non-coding (intron or promoter) regions of the genes screened; or there were technical limitations in the method of direct DNA sequencing. For example, the sequencing method used cannot detect copy number variants.
Given the heterogeneity in genetic etiologies and clinical manifestations of HCM, from asymptomatic to heart failure and sudden cardiac death, and even intra-family patients carrying the same gene mutation show impressively the wide spectrum of phenotypic presentation and outcome, some researchers questioned the appropriateness of attempts at genotype-phenotype correlation analyses [19]. Will the type of gene mutation really predict a certain clinical phenotype and the age of onset and prognosis of individual patients with HCM? Our study as well as the clues from human and animal model studies support that it is the number of mutation, not the type of mutation, predict the prognosis and disease course in HCM [12–16, 20].
At present, the risk markers of HCM has been established, including prior personal history of ventricular fibrillation, or sudden cardiac death, or sustained ventricular tachycardia, family history of sudden cardiac death, unexplained syncope episodes, non-sustained ventricular tachycardia, maximum left ventricular wall thickness ≥30 mm, abnormal blood pressure response during exercise. ICD placement is recommended for HCM patients with one of above-mentioned factors. However, most such factors have a low positive predictive value and the absence of risk factors does not convey absolute immunity to sudden cardiac death. The presence of >1 HCM-associated sarcomere mutation is associated with greater severity of disease, which provides an opportunity to predict clinical outcomes of HCM by using genetic information.
In our study, a positive correlation between the number of mutations and left ventricular hypertrophy and chamber enlargement of HCM was observed, supporting that the multiple gene mutations may be used to predict some clinical manifestations and prognosis of HCM. To some extent, MWT is not an ideal marker to predict clinical outcome. By following up, the correlation of mutations with the cardiac events can be better verified in young asymptomatic HCM patients.
We determined that about one-fifth of mutation carrier harbored two or more mutations in merely eight sarcomere genes. The proportion of multiple gene mutation carriers is expected to be even higher when additional HCM-related genes are screened. Our story suggested that new mutation searching efforts should not be suspended until a comprehensive genotype completed.
Conclusion
Our results once again proved that multiple mutations may be more practical and useful for prediction of HCM prognosis. Multiple mutations are much more frequent than that in literature reported by using more advanced sequencing technology and screening more HCM-related genes. Our result support that it is the number of mutation, not the type of mutation (such as R403Q mutation in MHY7), is the prognosis predictor.
References
Maron BJ (2002) Hypertrophic cardiomyopathy: a systematic review. JAMA 287:1308–1320
Zou Y, Song L, Wang Z, Ma A, Liu T, Gu H et al (2004) Prevalence of idiopathic hypertrophic cardiomyopathy in China: a population-based echocardiographic analysis of 8080 adults. Am J Med 116:14–18
Marian AJ (2010) Hypertrophic cardiomyopathy: from genetics to treatment. Eur J Clin Invest 40:360–369
Konno T, Chang S, Seidman JG, Seidman CE (2010) Genetics of hypertrophic cardiomyopathy. Curr Opin Cardiol 25(3):205–209
Williams JR (2008) The Declaration of Helsinki and public health. Bull World Health Organ 86:650–651
McKenna WJ, Spirito P, Desnos M, Dubourg O, Komajda M (1997) Experience from clinical genetics in hypertrophic cardiomyopathy: proposal for new diagnostic criteria in adult members of affected families. Heart 77:130–132
Simpson DA, Clark GR, Alexander S, Silvestri G, Willoughby CE (2011) Molecular diagnosis for heterogeneous genetic diseases with targeted high-throughput DNA sequencing applied to retinitis pigmentosa. J Med Genet 48(3):145–151
Jordan DM, Kiezun A, Baxter SM, Agarwala V, Green RC, Murray MF et al (2011) Development and validation of a computational method for assessment of missense variants in hypertrophic cardiomyopathy. Am J Hum Genet 88(2):183–192
Van Driest SL, Ellsworth EG, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ (2003) Prevalence and spectrum of thin filament mutations in an outpatient referral population with hypertrophic cardiomyopathy. Circulation 108:445–451
Olivotto I, Girolami F, Ackerman MJ, Nistri S, Bos JM, Zachara E et al (2008) Myofilament protein gene mutation screening and outcome of patients with hypertrophic cardiomyopathy. Mayo Clin Proc 83:630–638
Millat G, Bouvagnet P, Chevalier P, Dauphin C, Jouk PS, Da Costa A et al (2010) Prevalence and spectrum of mutations in a cohort of 192 unrelated patients with hypertrophic cardiomyopathy. Eur J Med Genet 53:261–267
Van Driest SL, Vasile VC, Ommen SR, Will ML, Tajik AJ, Gersh BJ et al (2004) Myosin binding protein C mutations and compound heterozygosity in hypertrophic cardiomyopathy. J Am Coll Cardiol 44:1903–1910
Richard P, Charron P, Carrier L, Ledeuil C, Cheav T, Pichereau C et al (2003) Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation 107:2227–2232
Richard P, Isnard R, Carrier L, Dubourg O, Donatien Y, Mathieu B et al (1999) Double heterozygosity for mutations in the b-myosin heavy chain and in the cardiac myosin binding protein C genes in a family with hypertrophic cardiomyopathy. J Med Genet 36:542–545
Ingles J, Doolan A, Chiu C, Seidman J, Seidman C, Semsarian C (2005) Compound and double mutations in patients with hypertrophic cardiomyopathy: implications for genetic testing and counselling. J Med Genet 42:e59
Girolami F, Ho CY, Semsarian C, Baldi M, Will ML, Baldini K et al (2010) Clinical features and outcome of hypertrophic cardiomyopathy associated with triple sarcomere protein gene mutations. J Am Coll Cardiol 55:1444–1453
Otsuka H, Arimura T, Abe T, Kawai H, Aizawa Y, Kubo T et al (2012) Prevalence and distribution of sarcomeric gene mutations in Japanese patients with familial hypertrophic cardiomyopathy. Circ J 76(2):453–461
Morita H, Rehm HL, Menesses A, McDonough B, Roberts AE, Kucherlapati R et al (2008) Shared genetic causes of cardiac hypertrophy in children and adults. N Engl J Med 358:1899–1908
Landstrom AP, Ackerman MJ (2010) Mutation type is not clinically useful in predicting prognosis in hypertrophic cardiomyopathy. Circulation 122:2441–2449
Tsoutsman T, Kelly M, Ng DC, Tan JE, Tu E, Lam L et al (2008) Severe heart failure and early mortality in a double-mutation mouse model of familial hypertrophic cardiomyopathy. Circulation 117:1820–1831
Acknowledgments
This study was supported by the Ministry of Science and Technology of China (grant number 2007DFC30340 and 2009DFB30050) and by the National Natural Science Foundation of China (grant number 30971233). We thank the patients for participating in the study. We are grateful to Ferhaan Ahmad for critical reading and helpful ideas of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Yubao Zou, Jizheng Wang, Xuan Liu, Lei Song and Rutai Hui contributed equally to the study
Rights and permissions
About this article
Cite this article
Zou, Y., Wang, J., Liu, X. et al. Multiple gene mutations, not the type of mutation, are the modifier of left ventricle hypertrophy in patients with hypertrophic cardiomyopathy. Mol Biol Rep 40, 3969–3976 (2013). https://doi.org/10.1007/s11033-012-2474-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-2474-2