Abstract
Aberrant expression of miRNAs plays critical roles in cancer development. Single nucleotide polymorphism (SNP) in miRNA precursors may affect miRNA expression levels. An important SNP in the pre-mir-27a with a A to G change (rs895819) was identified. Several original studies have explored the role of this SNP in cancer risk, but the results of these studies remain conflicting rather than conclusive. Therefore, we performed a meta-analysis of the published studies to derive a more precise estimation of the association between pre-mir-27a rs895819 polymorphism and cancer risk. In this meta-analysis, a total of 6 case–control studies (including 3,255 cases and 4,181 controls) were analyzed. The results of the overall meta-analysis did not suggest any associations between pre-mir-27a rs895819 polymorphism and cancer susceptibility. However, an decreased risk was observed in the subgroup of breast cancer patients (G vs A: OR = 0.90, 95 % CI = 0.83 ~ 0.97; P heterogeneity = 0.75) or in the subgroup of Caucasian race (G vs A: OR = 0.90, 95 % CI = 0.83 ~ 0.97, P heterogeneity = 0.78, I 2 = 0; AG vs AA: OR = 0.84, 95 % CI = 0.75 ~ 0.94, P heterogeneity = 0.35, I 2 = 3.7 %; GG+AG vs AA: OR = 0.85, 95 % CI = 0.76 ~ 0.94, P heterogeneity = 0.48, I 2 = 0). The findings suggest that pre-mir-27a rs895819 polymorphism may have some relation to breast cancer susceptibility or cancer development in Caucasian.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cancer is a major public health problem all over the world. In the United States, one in 4 deaths is due to cancer [1]. Currently, complete surgical resection is the effective treatment for this devastating disease. Early diagnosis is obviously important because it can increase the chance of a cure in early stage patients or prolong their survival time. However, most cancers at an early stage are difficult to detect. New markers for identifying high-risk populations as well as novel strategies for early detection are urgently needed. Now, mechanism of carcinogenesis is poorly understood. It has been suggested that susceptibility genes combining with environmental factors may be important in the development of cancer [2].
MicroRNAs (miRNAs, miRs) are a group of short non-coding, single-stranded RNAs, 18–25 nucleotides in length, which post-transcriptionally inhibit gene expression by either degrading or blocking translation of messenger RNA (mRNA) targets [3]. Currently, 2,042 homo sapiens mature miRNAs have been identified (The miRBase Sequence Database-Release 19.0) that can regulate most protein-coding genes [4]. It has been demonstrated that miRNAs are implicated in many important biological processes such as cell proliferation, differentiation, migration, apoptosis, and signal transduction, etc. [4, 5]. Aberrant expression of miRNAs play critical roles in carcinogenesis, too [6]. Single nucleotide polymorphism (SNP) in miRNA precursors may affect the expression of miRNAs. An important SNP in the pre-mir-27a with a A to G change (rs895819) was identified. Several original studies have explored the role of this SNP in cancer risk [7–12], but their results are conflicting rather than conclusive, which is partially attributed to the possible small effect of the SNP on cancer risk and/or the relatively small sample size in each of published studies. Therefore, we performed a meta-analysis of the published studies to derive a more precise estimation of the association between pre-mir-27a rs895819 polymorphism and cancer risk.
Materials and Methods
Publication search
In order to identify all the previous published studies on the association of pre-mir-27a rs895819 polymorphism with cancer, PubMed database was searched with the following keywords and subject terms: “rs895819” or “mir-27a” and “Single nucleotide polymorphism or SNP” by two independent investigators (last search update: September 19, 2012). The search was limited to the papers published in the English language. Reference lists were examined manually to further identify potentially relevant studies. All the studies matching the eligible criteria listed below were included in our meta-analysis.
Inclusion criteria
The following inclusion criteria were used in selecting literature for further meta-analysis: (1) evaluation of the pre-mir-27a rs895819 polymorphism and cancer risk; (2) a case–control design; (3) sufficient published data for calculating odds ratios (ORs) with their 95 % confidence intervals (95 % CIs).
Data extraction
Two investigators independently extracted the data. Discrepancies were adjudicated by the third investigator until consensus was achieved on every item. From each of the included articles the following information was abstracted: the name of first author, year of publication, country origin, ethnicity, cancer type, source of controls, genotyping methods, total number of cases and controls, the number of cases and controls with pre-mir-27a rs895819 polymorphism genotypes, G allele frequency in controls and P value for Hardy–Weinberg equilibrium (HWE), respectively.
Statistical methods
For the controls of each study, HWE was assessed using the Chi square goodness-of-fit test and a P < 0.05 was considered representative of a departure from HWE. The OR and its 95 % confidence interval (95 % CI) were used to assess the strength of the association between the pre-mir-27a polymorphism and cancer risk. The pooled ORs were performed for allelic comparison (G vs A), dominant model (GG+AG vs AA), recessive model (GG vs AG+AA), homozygote comparison (GG vs AA), and heterozygote comparison (AG vs AA), respectively. The statistical significance of the pooled ORs was determined by Z test, and P < 0.05 was considered statistically significant. A Chi square-based Q test was performed to assess the Inter-study heterogeneity. The fixed effect model (the Mantel–Haenszel method) [13] was used to access the pooled ORs if the heterogeneity was not significant (P > 0.1); otherwise, the random effect model (the DerSimonian and Laird method) [14] was used. Subgroup analyses were performed by ethnicity (categorized as Asian and Caucasian descents) and cancer type (categorized as breast cancer and gastrointestinal cancer). In addition, sensitivity analyses were performed to reflect the influence of the individual data on the summary ORs. Finally, Publication bias was evaluated using the Begg’s funnel plot and Egger’s test [15]. All statistical analyses were done with Stata software (Version 11; Stata Corporation, College Station, Texas, USA), and all tests were two-sided.
Results
Characteristics of the studies
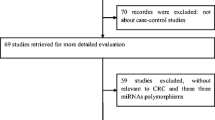
A total of 11 articles in English were achieved by an extensive search. We excluded five studies (two did not focus on rs895819, two were case-only studies, and one was not a case–control study). Finally, a total of 6 case–control studies met our inclusion criteria [7–12], including 3,255 cases and 4,181 controls. Table 1 presented the main characteristics of each study in the meta-analysis. There were three studies of Caucasian and three studies of Asian. Genotyping methods included DNA sequencing [7], polymerase chain reaction restriction fragment length polymorphism (PCR–RFLP) [8, 11], TaqMan [9, 10], and MassARRAY [12]. The genotype distributions in the controls of all studies were consistent with HWE.
Evidence synthesis
Of 4,181 control subjects included in this meta-analysis, 3,221 were Caucasians and 960 were Asians. The G allele frequencies for Caucasians and Asians were 32.55 % (95 % CI = 26.5 ~ 38.6 %) and 35.8 % (95 % CI = 11.6–60.0 %; P = 0.63), respectively. We observed a wide variation of G allele frequencies in Asian populations (Fig 1).
The results of the overall meta-analysis did not suggest any associations between rs895819 polymorphism and cancer susceptibility for all genetic models (Fig 2) (G vs A: OR = 0.95, 95 % CI = 0.84 ~ 1.09, P heterpgeneity = 0.02, I 2 = 64.3 %; GG vs AA: OR = 0.89, 95 % CI = 0.65 ~ 1.21, P heterpgeneity = 0.01, I 2 = 65.5 %; AG vs AA: OR = 1.02, 95 % CI = 0.83 ~ 1.26, P heterpgeneity < 0.01, I 2 = 72.4 %; AG+GG vs AA: OR = 0.99, 95 % CI = 0.82 ~ 1.20, P heterpgeneity = 0.01, I 2 = 70.2 %; and GG vs AG+AA: OR = 0.86, 95 % CI = 0.64 ~ 1.15, P heterpgeneity = 0.01, I 2 = 65.8 %).
Subgroup analyses were performed by cancer type, and the results showed that the rs895819 G allele was associated with decreased risk in breast cancer (G vs A: OR = 0.90, 95 % CI = 0.83 ~ 0.97; P heterpgeneity = 0.75), but same result was not presented in gastrointestinal cancer. In the stratified analysis by ethnicity, decreased risks were found in Caucasian in three of all genetic models tested (G vs A: OR = 0.90, 95 % CI = 0.83 ~ 0.97, P heterpgeneity = 0.78, I 2 = 0; AG vs AA: OR = 0.84, 95 % CI = 0.75 ~ 0.94, P heterpgeneity = 0.35, I 2 = 3.7 %; GG+AG vs AA: OR = 0.85, 95 % CI = 0.76 ~ 0.94, P heterpgeneity = 0.48, I 2 = 0). However, no significant association was observed in Asian.
Publication bias
Begger’s funnel plot and Egger’s test were used to assess the publication bias of included studies. The graphical funnel plots for all genetic models appeared to be symmetrical. Then, Egger’s test was used to provide statistical evidence of funnel plot symmetry. The results still did not show any evidence of publication bias in the overall meta-analysis (G vs A: t = 0.85, P = 0.45; GG vs AA: t = –0.37 P = 0.73; AG vs AA: t = 2.59 P = 0.06; GG+AG vs AA: t = 1.97 P = 0.12; GG vs AG+AA: t = –1.04 P = 0.36).
Sensitivity analysis
From the results of the leave-one-out sensitivity analysis, the study by Sun et al. could be the main cause of heterogeneity for allelic comparison (G vs A). After exclusion of this study, the heterogeneity no longer existed, and reached a positive association (G vs A: OR = 0.89, 95 % CI = 0.83 ~ 0.96, P heterpgeneity = 0.67, I 2 = 0 %) (Fig 3) (Table S1).
The influence of individual studies on the overall odds ratio (OR). The middle vertical axis indicated the overall OR and the two vertical axes indicated its 95 % confidence interval (CI). Every hollow round indicated the pooled OR when the left study was omitted in this meta-analysis. The two ends of every broken line represented the 95 % CI
Discussion
Much research effort has been directed toward understanding the role of mutations/SNPs present in precursor and mature miRNA and their influences on susceptibility of various diseases. Some studies have unambiguously demonstrated that sequence variations in miRNA precursors (pre-miRNA) can affect miRNA expression levels or function by interfering with the miRNA maturation process and/or binding activity to their mRNA targets, including rs895819 which is located in the loop of pre-miR-27a. Furthermore, differences in genotype distribution of this SNP have been found in a few case–control studies. To better understanding of the association between rs895819 polymorphism and cancer risk, we carried out a meta-analysis with a larger sample and subgroup analysis and to the best of our knowledge, this is the first review of the literature on their association.
Our results found no significant association between this polymorphism and cancer risk when all studies were pooled into the meta-analysis. If we removed the study by Sun et al. in the sensitive analysis, the heterogeneity no longer existed in allelic comparison, and reached a positive association, which suggested that this study might be the main source of heterogeneity for allelic comparison and this polymorphism would be associated with cancer risk if the sample size was large enough. Further analysis stratified by cancer type showed the rs895819 G allele was associated with decreased risk only in breast cancer (Table 2). Lack of significance in GI cancer subgroup might be due to the limited number of studies and the small sample size. In the other stratified analysis by ethnicity, the association between G allele of rs895819 and reduced risk of cancer was significant only in Caucasians. Besides the small sample size of Asian subgroup, the genetic diversity among ethnic groups might be another reason that no association was found. In our study, although G allele frequencies of Asians were similar to those of Caucasians, a wide variation of G allele frequencies was observed in Asian populations, which might be due to higher genetic diversity in Asian region and/or the limited subjects in the studies. These probably contributed to the heterogeneity of Asian subgroup in the racial analysis. In addition, Yang and Burwinkel [16] recently found that there might be a bias for genotyping of rs895819 by different genotyping methods due to the adjacent variant rs11671784, which is located only 4 nucleotides distance. Although Yang et al. used self-designed TaqMan allelic discrimination probes, which comprised only the target rs895819 site and avoided any overlap with the rs11671784, the TaqMan allelic discrimination assay still provided some false calls and introduced a bias. So they pointed out that the use of direct sequencing, restriction or MALDI-TOF Mass-ARRAY using primer extension from one direction (not overlapping with any SNP), was essential when investigating rs895819 and rs11671784, which deserved further study.
Recent studies found that miR-27a, as a oncomiR, exhibited its oncogenic activity through regulating target genes such as ZBTB10 [17–19], FOXO1 [20, 21], Spry2 [22], FBW7 [23–25], and PHB [26, 27] and so on. It means that down-regulation of miR-27a may contribute to decreased cancer risk through up-regulating the targets. Although the binding of the mature miRNA to target mRNAs was not influenced by the SNP rs895819, some studies had demonstrated that polymorphisms in pre-miRNAs could influence the expression of their mature forms, as well as were involved in the binding of some nuclear factors in miRNA processing [28, 29]. So we presumed that rs895819 affected the processing or expression of miR-27a, which resulted in down-regulation of miR-27a. The presumption was supported by our findings in breast cancer subgroup or Caucasian subgroup.
Our meta-analysis should be interpreted after considering some limitations. First, the effect of gene–gene and gene-environment interactions was not addressed in the analysis. Second, small study number could bias the results in subgroup analyses. In addition, it was also necessary to conduct larger sample studies using homogeneous cancer patients and well matched controls. Only based the well-designed studies with the above factors taken into account, a better, comprehensive understanding of the association between the pre-mir-27a A>G polymorphism and cancer risk is obtained.
References
Siegel R, Naishadham D, Jemal A (2012) Cancer statistics, 2012. CA Cancer J Clin 62:10–29
Lichtenstein P et al (2000) Environmental and heritable factors in the causation of cancer–analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med 343:78–85
He L, Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5:522–531
Anglicheau D, Muthukumar T, Suthanthiran M (2010) MicroRNAs: small RNAs with big effects. Transplantation 90:105–112
Kutanzi KR et al (2011) MicroRNA-mediated drug resistance in breast cancer. Clin Epigenetics 2:171–185
Esquela-Kerscher A, Slack FJ (2006) Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer 6:259–269
Yang R et al (2009) A genetic variant in the pre-miR-27a oncogene is associated with a reduced familial breast cancer risk. Breast Cancer Res Treat 121:693–702
Sun Q et al (2010) Hsa-mir-27a genetic variant contributes to gastric cancer susceptibility through affecting miR-27a and target gene expression. Cancer Sci 101:2241–2247
Catucci I et al (2012) The SNP rs895819 in miR-27a is not associated with familial breast cancer risk in Italians. Breast Cancer Res Treat 133:805–807
Hezova R et al (2012) Evaluation of SNPs in miR-196-a2, miR-27a and miR-146a as risk factors of colorectal cancer. World J Gastroenterol 18:2827–2831
Zhang M et al (2012) Associations of miRNA polymorphisms and female physiological characteristics with breast cancer risk in Chinese population. Eur J Cancer Care (Engl) 21:274–280
Zhou Y et al (2012) Association analysis of genetic variants in microRNA networks and gastric cancer risk in a Chinese Han population. J Cancer Res Clin Oncol 138:939–945
Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22:719–748
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7:177–188
Egger M et al (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315:629–634
Yang R, Burwinkel B (2012) A bias in genotyping the miR-27a rs895819 and rs11671784 variants. Breast Cancer Res Treat 134:899–901
Mertens-Talcott SU et al (2007) The oncogenic microRNA-27a targets genes that regulate specificity protein transcription factors and the G2-M checkpoint in MDA-MB-231 breast cancer cells. Cancer Res 67:11001–11011
Li X et al (2010) MicroRNA-27a indirectly regulates estrogen receptor alpha expression and hormone responsiveness in MCF-7 breast cancer cells. Endocrinology 151:2462–2473
Chintharlapalli S et al (2011) Betulinic acid inhibits colon cancer cell and tumor growth and induces proteasome-dependent and -independent downregulation of specificity proteins (Sp) transcription factors. BMC Cancer 11:371
Guttilla IK, White BA (2009) Coordinate regulation of FOXO1 by miR-27a, miR-96, and miR-182 in breast cancer cells. J Biol Chem 284:23204–23216
Zhou L et al (2012) Mechanism and function of decreased FOXO1 in renal cell carcinoma. J Surg Oncol 105:841–847
Ma Y et al (2010) miR-27a regulates the growth, colony formation and migration of pancreatic cancer cells by targeting Sprouty2. Cancer Lett 298:150–158
Lerner M et al (2011) MiRNA-27a controls FBW7/hCDC4-dependent cyclin E degradation and cell cycle progression. Cell Cycle 10:2172–2183
Spruck C (2011) miR-27a regulation of SCF(Fbw7) in cell division control and cancer. Cell Cycle 10:3232–3233
Wang Q et al (2011) Upregulation of miR-27a contributes to the malignant transformation of human bronchial epithelial cells induced by SV40 small T antigen. Oncogene 30:3875–3886
Fletcher CE et al (2012) Androgen-regulated processing of the oncomir MiR-27a, which targets prohibitin in prostate cancer. Hum Mol Genet 21:3112–3127
Liu T et al (2009) MicroRNA-27a functions as an oncogene in gastric adenocarcinoma by targeting prohibitin. Cancer Lett 273:233–242
Jazdzewski K et al (2008) Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci USA 105:7269–7274
Duan R, Pak C, Jin P (2007) Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum Mol Genet 16:1124–1131
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81272470).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhong, S., Chen, Z., Xu, J. et al. Pre-mir-27a rs895819 polymorphism and cancer risk: a meta-analysis. Mol Biol Rep 40, 3181–3186 (2013). https://doi.org/10.1007/s11033-012-2392-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-2392-3