Abstract
The mitogen-activated protein (MAP) kinase cascade is an important signaling module which is involved in biotic and abiotic stress responses as well as plant growth and development. In this study, we identified 17 tobacco MAPKs including 11 novel tobacco MAPK genes that have not been identified before. Comparative analysis with MAPK gene families from other plants, such as Athaliana thaliana, rice and poplar, suggested that tobacco MAPKs (such as NtMPK1, NtMPK3 and NtMPK8) might play similar functions in response to abiotic and biotic stresses. QRT-PCR analysis revealed that a total of 14 NtMPKs were regulated by SA and/or MeJA, suggesting their potential roles involved in plant defense response. In addition, 6 NtMPKs were induced by drought treatment, implying their roles in response to drought stress. Our results indicated that most of tobacco MAPK might be involved in plant defense response, which provides the basis for further analysis on physiological functions of tobacco MAPKs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The mitogen-activated protein kinase cascade is an important signaling module that transduces exogenesis or endogenesis signaling molecules to intracellular responses in eukaryotic organisms. Typically, the conserved signaling module consists of three kinds of protein kinases: MAPK kinase kinase (MAP3 K), MAPK kinase (MKK) and MAPK [1]. As the terminal component of this cascade, MAPKs are activated through phosphorylation on their threonine and tyrosine residues by their upstream MAP2 K and MAP3 K. At the downstream of this cascade, some transcription factors and other signaling components [2, 3], which have important roles in plant immunity through activation of downstream genes, serve as the physiological substrates of MAP kinases.
For recent decades years, mitogen-activated protein kinase cascade drew much attention not only because its crucial roles in responding to various extracellular stimuli [4–8] but also its roles in severing as a universal signaling pathway which regulates various cellular activities, such as cell division, differentiation and programmed cell death [6, 9, 10]. In Arabidopsis, the flagellin flg22 may activate a MAPK cascade including MEKK1, MKK4/MKK5 and MPK3/MPK6, which, in turn, increase the expression of WRKY29 and confer resistance to P. syringae (Ps) as well as B. cinerea [2]. Interestingly, isolation of another MAPK from Arabidopsis, AtMPK4, showed its role to negatively regulate systemic acquired resistance [11]. Alfalfa MAP kinase 3 (MMK3) could be found during all stages of the cell cycle, but its protein kinase activity was only detected in mitosis and correlated with the timing of phragmoplast formation, suggesting its roles in the regulation of plant cytokinesis [10]. Besides, increasing evidence reveals that MAP kinases are involved in plant priming mechanism and induced-resistance (IR) phenomena, which suggests plants have the capacity of “remembering” and responding to environment stimuli in a more rapid and robust manner [12, 13]. A typical research on Arabidopsis revealed that prestress deposition of the signaling components MPK3 and MPK6 was a critical step in priming plants for full induction of defense responses during IR [14].
Based on the genome scan of several plant genomes, MAPK families have been identified in various plants. For example, A. thaliana genome possesses genes encoding 23 MAPKs [15, 16] compared with 17 MAPKs in rice [8] and 21 MAPK homologs in poplar [17]. Interestingly, as a dicot plant, grapevine possesses only 12 MAPK genes [18], which is very similar with human [19] and zebrafish [20] (both has 10 MAPKs respectively). However, there is no such study on analysis of tobacco MAPK gene family on the whole genome. According to our knowledge, only six tobacco MAPKs (SIPK [21], WIPK [22, 23], Ntf4 [24], NRK1 [25], Ntf3 [26] and MPK4 [27]) were analyzed. The salicylic acid-inducible protein kinase (SIPK) and the wound-inducible protein kinase (WIPK), which are the first two isolated MAPKs from tobacco, were proved to be activated in response to various environmental stresses [21, 28–31]. Then, another study showed that suppression of NtSIPK and NtWIPK attenuated the N gene-mediated resistance against tobacco mosaic virus (TMV), which suggests that these two MAPK were required for TMV defense [32]. Another two MAPKs, Ntf4 and NRK1, were reported to be involved in regulating pathogen-induced HR cell death in tobacco [24, 25, 29]. In addition to the function of plant resistance, other two MAPKs (Ntf4 and NRK1) were considered as positive regulators of plant development and cell cytokinesis [33–35]. Loss-of-function experiment of NtMPK4 revealed that NtMPK4 had played important roles in JA signaling and stomatal movement [27].
In this study, we firstly provided a list of MAPK members from tobacco. Based on cloning and sequencing, we got 17 MAPKs and 11 of 17 MAPK genes were novel. We classified these 17 MAPKs into six groups according to the phylogenetic analysis. Besides, we investigated the conserved domains and divergent sequences among the 17 MAPK by multi alignment of protein sequences. Further investigation revealed that some tobacco MAPK genes could be regulated by methyl jasmonate acid (MeJA), salicylic acid (SA) and drought using quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis. Our data provide some new insights into potential functions of the tobacco MAPKs.
Materials and methods
Plant materials and preparation of RNA and first strand cDNA synthesis
Common tobacco (Nicotiana tabacum cv HHDJY) seeds sterilized by 10 % NaClO were germinated in sterilized planets with MS solid culture medium in a green house programmed for 22 °C and a 14-h light cycle. After 8 weeks, tobacco plants were treated by phytohormone as followed: for SA, and MeJA treatments, tobacco seedlings were cultured in solutions with 2 mM SA in water, and 100 μM JA in 0.01 % ethanol, respectively [36]. For drought treatment, tobacco seedlings were placed sterilized filter paper without any water.
TRIpure reagent (Roche, China) was used to extract total RNA from various treated tobacco seedlings according to the manufacturer’s protocol. RT-PCR was completed to synthesize the first-strand cDNA with 2 μg of purified total RNA (pre-treated with DNase I) according to the manufacturer’s protocol from Promega. The oligo (dT)18 was used as a primer and the reverse transcription reaction was incubated at 42 °C for 65 min in a total volume of 26 μl. The reverse transcription products were diluted to 100 μl with sterile ddH2O.
Cloning and identification of MAPKs from tobacco
Several sources, such as ESTs, TGI (Tobacco Genome Initiative, http://www.pngg.org/tgi/) and N. benthamiana genome (http://solgenomics.net/organism/Nicotiana_benthamiana/genome) are available to provide bioinformatics analysis of tobacco MAPKs. Using these released data, 13 primer pairs were designed using PrimerPremier 5.0 (Supplementary Table 1) to amplify the full-length open reading frame of tobacco MAPK genes. Each polymerase chain reaction (PCR) contained a total volume of 50 μl, containing 10 ng cDNA template, 0.2 μM of each primer and 25 μl GoTaq Green mix (Promega, USA). The PCR conditions included 4 min initial denaturation at 94 °C, followed by 33 cycles of the following: 30 s denaturation at 94 °C, 45 s annealing at appropriate temperature, 2 min of extension at 72 °C and a final 10 min extension at 72 °C. The amplified products were purified with agarose gel purification kit (Bioteck, China) and then cloned into pEASY-T1 vector (Transgen, China). All positive clones were sequenced by BGI Company. The raw reads were assembled using BioEdit software.
To identify novel tobacco MAPKs, we followed the criteria suggested by Nathan [8]: (i) their amino acid sequences are significantly homologous with other known MAPKs; (ii) there are 11 conserved subdomains in the MAPKs kinase domain; (iii) the TDY or TEY motif is essential in the activation site.
Conserved domain detection
Protein secondary structure and domain provide much information for researchers to understand the function of these proteins. Therefore, the multiple sequence alignment of the amino acid sequences and kinase domains of MAPKs was performed using the MultAlin program [37].
Phylogenetic analysis
The amino acids sequences of MAPKs from four plants (Arabidopsis, tobacco, rice, poplar) were initially aligned using Clustal W (v.1.83) [38]. The phylogenetic tree for the plants MAPKs was reconstructed by Neighbor-Joining method in which distance was estimated by JTT amino acid matrix implemented in the program MEGA 4.0 [39]. The pairwise deletion option was set in the NJ tree reconstruction and the accuracy of the tree topology was assessed by bootstrap analysis with 1,000 resampling replicates.
Quantitative real-time PCR
A pair of gene-specific primers was designed by PrimerPremier 5.0 according to tobacco MAPK gene sequences (Supplementary Table 2). In order to avoid the effect of potential genomic DNA contamination, these primers were designed to span intron-splicing sites if possible. Tobacco EF-1α gene (Genbank: D63396.1) was used as a control. Tobacco PR1a and ODC were analyzed to verify the accuracy of SA and MeJA treatment, respectively. The real-time PCR analysis were performed using BIO-RAD CFX96 real-time PCR system (Bio-Rad, USA 96 well) formats with programs as following: an initial 95 °C denaturation step for 3 min, followed by 40 cycles of denaturation at 95 °C for 10 s and annealing/extension at 60 °C for 1 min. The disassociation curve was used to detect the effect of amplification. Each PCR reaction was mixed with 7.5 μl of SYBR Green (Takara, China), 4.5 μl of ddH2O, 1 μl of each reverse-transcribed and digested cDNA product, and 1 μl of each primer (10 μM).
Relative gene expression level was analyzed according to 2−ΔΔCT method. The ΔCT and ΔΔCT were calculated by the formulas ΔCT = CT target−CT control and ΔΔCT = ΔCT treated sample−ΔCT untreated sample (0 h treatment), respectively. In our study, genes with only threefold change compared with 0 h control were considered induced.
Results
Identification and nomenclature of the tobacco MAPK genes
As we have mentioned above, six tobacco MAPK genes were reported previously and most of them were involved in environmental stimuli as well as plant development and cell cytokinesis. However, other members of tobacco MAPK gene family have not been identified and characterized. After extensive in silico analysis of the released data from tobacco EST and TGI, we cloned 13 novel MAPK homologs. However, according to the criteria proposed by Nathan [8], two of them were not considered as MAPK gene family members since the first two conserved subdomains did not exist in their protein kinase domains (data not shown), despite of their significant homology with other tobacco MAPKs at the amino acid level. Thus, in total, we got 17 tobacco MAPKs and 11 of 17 were novel which were not identified before. To avoid confusing names, these 17 tobacco MAPKs (Table 1) were named according to the nomenclature followed by previous study in Arabidopsis [16].
Phylogenetic analysis of tobacco MAPK genes
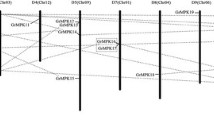
In order to evaluate the evolutionary relationship of MAPKs between tobacco and other plants, and predict the potential orthologous and paralogous relations among MAPKs in the four model plants, a combined phylogenetic tree (Fig. 1) was constructed using software MEGA 4.0 with NJ method. The results showed that MAPK proteins could be divided into six major groups (Group A–F), and each group contains at least two tobacco MAPKs. According to the difference of TEY or TDY motif in their activation site, all the six groups can be divided into two subtypes. Groups A, B, C and F contains TEY motif, whereas TDY motif exists in group D and E. We investigated the numbers of MAPK belonging to the TEY type as well as TDY type from the four species. The results showed that the members belonging to TEY were more than members belonging to TDY in dicot plants, whereas in monocots plants, this situation would be different. For example, we found 15 AtMPKs, 12 PtMPKs and 10 NtMPKs belonged to TEY motif whereas only 8 AtMPKs, 9 PtMPKs and 6 NtMPKs belonged to TDY. However, in rice, 5 OsMPKs contained the TEY motif whereas 11 OsMPKs contained TDY motif. Those results suggested that MAPKs containing the TEY motif might play more important roles than MAPKs belonging to the TDY motif in dicot plants. Interestingly, not all of the MAPK family members contain the TEY or TDY motif in their kinase activation loop. A MEY motif was also found in both OsMPK2 [40] and NtMPK2 whereas none was found in Arabidopsis and poplar MAPK genes.
Phylogenetic relationships of MAPK gene families from tobacco, A. thaliana, rice and poplar. Amino acid sequences were aligned using ClustalX and subjected to phylogenetic analysis using the NJ method with 1,000 resampling replicates. The red asterisks in the branches indicate potential orthologous/paralogous relationship
Group A contains the most extensively studied plant MAPKs, such as AtMPK3/6, OsMPK1/5 and NtMPK5/6 (known as WIPK/SIPK). Most of MAPKs in this group have been reported to be associated with a variety of biotic and abiotic stress responses. For example, NtMPK7 (known as Ntf4) has been illustrated to be involved in plant defense response [24] and a possible relationship to the gametophytic function during plant development [35]. Group B contains four NtMPKs (NtMPK1, NtMPK2, NtMPK3 and NtMPK4). All of the four NtMAPKs share high similarity with OsMPK6 and AtMPK4, both of which are reported to regulate negatively systemic acquired resistance [11, 41]. Group C includes NtMPK14 and NtMPK15. NtMPK14 share higher similarity with OsMPK4, a gene proved to be induced by wounding, JA, SA, ABA, ET, H2O2, salt in rice [7]. While NtMPK15 has high identities with AtMPK7 whose circadian-rhythm-regulated expression was detected by microarray analysis.
Group D and E, which are notable for their TDY motif in the activation site and the extended C-terminal sequences, include a total of six NtMPKs (for Group D: NtMPK8/9; for Group E: NtMPK10-13). However, only one of them, NtMPK8 (known as Ntf3) [26], was isolated and studied.
Group F contained the least members of plant MAPKs. Only two NtMPKs (NtMPK16 and NtMPK17) were found in this branch. To date, few of work have been done to analyze the function of MAPKs in this group.
Multiple sequence alignment and structure analysis
To explore the conserved and diverse function of tobacco MAPKs, multiple sequence alignment was performed with the deduced kinase domains (Fig. 2). As reported previously, all tobacco MAPKs proteins contained 11 highly conversed kinase subdomains as well as TXY motif (except NtMAPK2), which was considered as the typical characteristic of MAPK family. It was suggested that these MAPK domains should play important roles in plants.
Protein sequence alignment of tobacco MAPKs kinase domain. Alignment was performed using the MultAlin 5.4.1 program. The high-, low-, and neutral- consensus amino acid (aa) residues are depicted in red, blue and black colors, respectively. Highly conserved aa residues appear in high-consensus color and as an uppercase letter in the consensus line. The PK subdomains are in italicized roman numerals (I–XI) on the top of each row and aa positions are marked by green lines. The phosphorylation-activation motif (TEY and TDY) is indicated by yellow box
In addition to the conserved domains, divergent sequences were also detected in the regions between subdomain VII and VIII (Fig. 2), although we are not familiar with their functions. Previous research demonstrated that SIPK (NtMPK6) contained a common docking (CD) domain (LNSLHDISDEPICM) at the C-teminus, which is involved in the interaction with their upstream kinases, substrates and negative regulator including MAPK phosphatases [5]. In our analysis, CD domains were only observed in members of Group A, B and C (Supplementary Fig. 1), indicating these genes may have similar functions. Group D, E and F have quite divergent C-terminal extension (Supplementary Fig. 1) suggesting that they might have diverse functions in plant cellular activities.
Expression profiles of NtMAPKs in response to various signals
Increasing evidence showed that phytohormone SA and MeJA play diverse and important roles in plant defense response [42]. To explore the function of tobacco MAPKs in response to these defense signaling molecules, we analyzed the expression patterns of tobacco MAPKs under treatment of SA and MeJA (Fig. 3a, b). Nine tobacco MAPK genes have been regulated by SA (Fig. 3a). Among them, the expression of five NtMPKs (NtMPK7/8/10/11/13) was up-regulated under SA treatment, with NtMPK11 and NtMPK13 to be considered highly induced at 10 h treatment by SA. The remaining four MAPKs (NtMPK3/15/16/17) showed down-regulated expression under SA treatment, exceptionally NtMPK15. In our MeJA treatment experiment, 11 MAPKs (NtMPK1/4/5/6/7/8/10/14/15/16/17) had inducible expression. Among them, 10 genes showed up-regulated expression, especially NtMPK5 and NtMPK16, which were up-regulated 40-folds higher than control at 10 h of treatment. And only one gene, NtMPK4, was slightly down-regulated by MeJA. Our results indicated most of tobacco MAPKs were involved in response to SA and MeJA signaling pathway and might have important functions in plant defense response.
Many studies demonstrated that MAPKs involved in response to drought stress [5, 43, 44]. For example, the transcript levels of an alfalfa MAPK gene MMK4 [43] and GhMPK16 [44] accumulated after drought treatment. In our qRT-PCR analysis, the mRNA levels of six tobacco MAPKs (NtMPK1/3/9/10/14/15) were induced by drought treatment. NtMPK1 and NtMPK9 were significantly induced by drought treatment (Fig. 3c). The results suggested several tobacco MAPKs might be involve in response to drought stress.
Discussion
A list of 17 tobacco MAPKs were firstly analyzed in our study. And among them, we cloned 13 novel MAPK homologs in common tobacco, among which 2 genes were excluded according to the criteria proposed by Nathan [8]. Although N. tabacum has a very large genome size with 4.5 billion base pairs compared with other plants, mobile DNAs may make up the majority of the nuclear genome [45]. Therefore, considering the conserved numbers of MAPKs from other plants (Arabidopsis: 23, rice: 17, poplar: 21), it is likely that we have identified most, if not all, of the MAPKs in N. tabacum.
Previous study in A. thaliana indicated that MAPK were clustered into four groups [16]. However, the phylogenetic tree in this study reveals that tobacco MAPKs appear to be clustered into six groups (Group A–F). Base on the phylogenetic analysis, several potential orthologous/paralogous subclusters were identified (Fig. 1) in our research. These potential orthologous relationships help us to understand possible functions of tobacco MAPKs. Previous research revealed that AtMPK4 was required for jasmonic acid-responsive gene expression and negatively regulated systemic acquired resistance in Arabidopsis [11]. We speculate its orthologs, NtMPK1 and NtMPK2, may share a similar function in plant defense response. Actually, Kenji has reported NtMPK1 (old name NtMPK4) is required for jasmonic acid signaling and involved in ozone tolerance via stomatal movement in tobacco [27]. Our qRT-PCR analysis (Fig. 3b) reveals NtMPK1 is induced by MeJA treatment, which is similar to Kenji’s results. Similarly, it has been investigated that OsMPK6 functioned both as an activator and a repressor to regulate rice defense responses [46]. As its ortholog, NtMPK3 was down-regulated by SA in our qRT-PCR analysis (Fig. 3a), which suggested NtMPK3 had a similar function with OsMPK6 in negatively regulating systemic acquired resistance (SAR). In addition, it is likely that NtMPK3 is also involved in response to drought stress since its transcript level is induced by drought treatment (Fig. 3c). Although no more information about NtMPKs in Group D is known, rice MAPKs in the group support more useful details to analyze NtMPKs. Two OsMPKs (OsMPK7 and OsMPK8) were found to be involved in biotic and abiotic stress responses in group D. Over-expression and suppression of OsMPK7 revealed that the MAPK participate in JA signaling and defense response [40]. While OsMPK8 could be regulated by ABA, H2O2, drought, temperature changing, heavy metals and UV–C irradiation, and was also associated with host cell death [8]. These studies suggest that NtMPK8 (also known as Ntf3), the ortholog of OsMPK7/8, may play an important role in response to various environmental stimuli and phytohormone signaling. Our qRT-PCR analysis partly verified the speculation since we detected up-regulated expression of NtMPK8 by SA and MeJA treatment (Fig. 3a, b); however, no obvious change was observed under drought treatment.
Salicylic acid (SA) and jasmonic acid (JA) are two important signaling molecules involved in plant defense against pathogen infection. Accumulation of SA is required for the induction of SAR [47]; while, JA is involved in induced systemic resistance (ISR) against pathogens [48]. MAPK induced by SA and/or JA may be involved in plant defense response against pathogens. OsMPK12 (also known as BWMK1) was induced by both SA and JA treatments in Nathan’s qRT-PCR analysis [8]. Constitutive expression of OsBWMK1 also enhanced resistance against pathogen infections [49]. We speculate tobacco MAPKs (NtMPK1/3/4/5/6/7/8/10/11/13/14/15/16/17) induced by SA and/or MeJA in our qRT-PCR analysis may play important roles in plant SAR and ISR. It has been reported that kinase activity of SIPK (NtMPK6) was activated by SA [21]. However, mRNA level of SIPK was not detected obviously induced by SA in our qRT-PCR analysis. To validate the accuracy and effect of our treatments with SA and MeJA, expression of PR1a [50] and ODC [51] in tobacco was analyzed as marker genes through qRT-PCR (Supplementary Fig. 2a, b), respectively. Our results showed that the treatments in our research were effective. This phenomenon suggests that those MAPKs, which are not induced by SA at mRNA level, may play important roles in plant defense response. Interestingly, both WIPK and SIPK were induced by MeJA at mRNA levels in qRT-PCR analysis (Fig. 3b). Actually, our analysis verified the fact that WIPK and SIPK were involved in JA production [52].
Our results will provide important information of tobacco MAPKs for further research. Detection of mRNA level change could not return real information sometimes. We expect transgenic analysis and kinase activity assay, which are effective ways to understand the functions of MAPKs, will be established in the future.
References
Christian W et al (1999) Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol Rev 79(1):143–180
Tsuneaki A et al (2002) MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415:977–983
Ishihama N et al (2011) Phosphorylation of the Nicotiana benthamiana WRKY8 transcription factor by MAPK functions in the defense response. Plant Cell Online 23(3):1153–1170
Hirt H (1997) Multiple roles of MAP kinases in plant signal transduction. Trends Plant Sci 2(1):131–144
Shuqun Z, Daniel FK (2001) MAPK cascades in plant defense signaling. Trends Plant Sci 6(11):502–527
Claudia J et al (2002) Complexity, cross talk and integration of plant MAP kinase signalling. Curr Opin Plant Biol 5:415–424
Agrawal GK et al (2003) Novel rice MAP kinases OsMSRMK3 and OsWJUMK1 involved in encountering diverse environmental stresses and developmental regulation. Biochem Biophys Res Commun 300(3):775–783
Nathan SR, Yinong Y (2006) Molecular analysis of the rice MAP kinase gene family in relation to Magnaporthe grisea infection. Mol Plant Microbe Interact 19:530–540
Calderini O et al (1998) A cell cycle regulated MAP kinase with a possible role in cytokinesis in tobacco cells. J Cell Sci 111(Pt 20):3091–3100
László B et al (1999) A MAP kinase is activated late in plant mitosis and becomes localized to the plane of cell division. Plant Cell 11:101–113
Morten P, Peter B, Henrik N (2000) Arabidopsis MAP kinase 4 negatively regulates systemic aquired resistance. Cell 103:1111–1120
Uwe C, Corné MJP, Brigitte M–M (2002) Priming in plant–pathogen interactions. Trends Plant Sci 7(5):210–216
Beckers GJM, Conrath U (2007) Priming for stress resistance: from the lab to the field. Curr Opin Plant Biol 10(4):425–431
Beckers GJM et al (2009) Mitogen-activated protein kinases 3 and 6 are required for full priming of stress responses in Arabidopsis thaliana. Plant Cell Online 21(3):944–953
Guillaume T et al (2001) Plant mitogen-activated protein kinase signaling cascades. Curr Opin Plant Biol 4:392–400
Ichimura Kazuya et al (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci 7:301–308
Hamel L-P et al (2006) Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci 11(4):192–198
Tae KH et al (2010) Comparative genomic analysis of mitogen activated protein kinase gene family in grapevine. Genes Genomics 32:275–281
Krens S, Spaink H, Snaarjagalska B (2006) Functions of the MAPK family in vertebrate-development. FEBS Lett 580(21):4984–4990
Krens SFG et al (2006) Characterization and expression patterns of the MAPK family in zebrafish. Gene Expr Patterns 6:1019–1026
Shuqun Z, Klessig DF (1997) Salicylic acid activates a 48-kD MAP kinase in tobacco. Plant Cell 9:809–824
Shigemi S, Hiroshi S, Ohashi Y (1999) Jasmonate-based wound signal transduction requires activation of WIPK, a tobacco mitogen-activated protein kinase. Plant Cell 11:289–298
Shuqun Z, Yidong L, Daniel FK (2000) Multiple levels of tobacco WIPK activation during the induction of cell death by fungal elicitins. Plant J 23(3):339–347
Dongtao R et al (2006) Activation of Ntf4, a tobacco mitogen-activated protein kinase, during plant defence response and its involvement in hypersensitive response-like cell death. Plant Physiol 141:1482–1493
Yule L, Michael S, Dinesh-Kumar SP (2004) Involvement of MEK1 MAPKK, NTF6 MAPK, WRKY/MYB transcription factors, COL1 and CTR1 in N-mediated resistance to tobacco mosaic virus. Plant J 38:800–809
Wilson C et al (1993) Isolation and characterization of a tobacco cDNA clone encoding a putative MAP kinase. Plant Mol Biol 23(3):543–551
Gomi K et al (2005) A mitogen-activated protein kinase NtMPK4 activated by SIPKK is required for jasmonic acid signaling and involved in ozone tolerance via stomatal movement in tobacco. Plant Cell Physiol 46(12):1902–1914
Dhirendra K, Daniel FK (1999) Differential induction of tobacco MAP kinases by the defense signals nitric oxide, salicylic acid, ethylene, and jasmonic acid. Mol Plant Microbe Interact 13:347–351
Kwang-Yeol Y, Yidong L, Shuqun Z (2000) Activation of a mitogen-activated protein kinase pathway is involved in disease resistance in tobacco. PNAS 98:741–746
Mary E, Hoyos, Shuqun Z (2000) calcium-independent activation of salicylic acid-induced protein kinase and a 40-kilodalton protein kinase by hyperosmotic stress. Plant Physiol 122:1355–1363
Monika M, Ajczyk et al (2000) osmotic stress induces rapid activation of a salicylic acid–induced protein kinase and a homolog of protein kinase ASK1 in tobacco cells. Plant Cell 12:165–178
Hailing J et al (2002) Function of a mitogen-activated protein kinase pathway in N gene-mediated resistance in tobacco. Plant J 33:719–731
Soyano T (2003) NQK1/NtMEK1 is a MAPKK that acts in the NPK1 MAPKKK-mediated MAPK cascade and is required for plant cytokinesis. Genes Dev 17(8):1055–1067
Michiko S et al (2006) Phosphorylation of NtMAP65-1 by a MAP kinase down-regulates its activity of microtubule bundling and stimulates progression of cytokinesis of tobacco cells. Genes Dev 20:1004–1014
Maria-Jose C et al (2007) In situ molecular identification of the Ntf4 MAPK expression sites in maturing and germinating pollen. Biol Cell 99:209–221
Gaiyun Z et al (2009) Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J Exp Bot 60(13):3781–3796
Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16(22):10881–10890
Higgins DG, Thompson JD, Gibson TJ (1996) Using CLUSTAL for multiple sequence alignments. Comput Methods Macromol Seq Anal 266:383–402
Tamura K et al (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24(8):1596–1599
Jai SR, Yinong Y (2007) Rice Mitogen-activated Protein Kinase Gene Family and Its Role in Biotic and Abiotic Stress Response. J Integr Plant Biol 49(6):751–759
Shen X, Yuan B, Liu H, Li X, Xu C, Wang S (2010) Opposite functions of a rice mitogen-activated protein kinase during the process of resistance against Xanthomonas oryzae. Plant J 64:86–99
Yang Y, Shah J, Klessig DF (1997) Signal perception and transduction in plant defense responses. Genes Dev 11(13):1621–1639
Claudia J et al (1996) Stress signaling in plants: a mitogen-acticated protein kinase pathway is acticated by cold and drought. Proc Natl Acad Sci USA 93:11274–11279
Shi J et al (2011) GhMPK16, a novel stress-responsive group D MAPK gene from cotton, is involved in disease resistance and drought sensitivity. BMC Mol Biol 12:22
Bennetzen JL (2000) Transposable element contributions to plant gene and genome evolution. Plant Mol Biol 42:251–269
Yuan B et al (2007) Mitogen-activated protein kinase OsMPK6 negatively regulates rice disease resistance to bacterial pathogens. Planta 226(4):953–960
Ryals John et al (1995) Signal transduction in systemic acquired resistance. Proc Natl Acad Sci USA 92:4202–4205
Heil Martin, Bostock RM (2002) Induced Systemic Resistance (ISR) Against Pathogens in the Context of Induced Plant Defences. Ann Bot 89:503–512
Koo SC et al (2009) OsBWMK1 mediates SA-dependent defense responses by activating the transcription factor OsWRKY33. Biochem Biophys Res Commun 387(2):365–370
Payne George et al (1989) Isolation and sequence of a genomic clone encoding the basic form of pathogenesis-related protein 1 from Nicotiana tabacum. Plant Mol Biol 12:595–596
Xu Bingfang, Sheehan MoiraJ, Timko MP (2004) Differential induction of ornithine decarboxylase (ODC) gene family members in transgenic tobacco (Nicotiana tabacum L. cv. Bright Yellow 2)cell suspensions by methyl-jasmonate treatment. Plant Growth Regul 44:101–116
Seo Shigemi et al (2007) The mitogen-activated protein kinases WIPK and SIPK regulate the levels of jasmonic and salicylic acids in wounded tobacco plants. Plant J 49:899–909
Acknowledgments
This work was supported by grants from the National Basic Research Program of China (No. 2012CB114600), Fundamental Research Funds for the Central Universities (No. CDJZR10290003), the National Natural Science Foundation (No. 30901054, 31001034 and 31000563) and Key Project for National Tobacco Government (No. 110200902037).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Full-length protein sequence alignment of tobacco MAPKs. The CDdomains were marked by a red box (GIF 97 kb)
Supplementary Fig. 2
Expression of PR1a and ODC under treatments of SA and MeJA (JPG 31 kb)
Supplementary Table 1
The primer sequences used in RT-PCR for cloning of 11 novol NtMPKs. (DOC 24 kb)
Supplementary Table 2
The primer sequences used in qRT-PCR for detection of NtMPK expression. (DOC 37 kb)
Rights and permissions
About this article
Cite this article
Zhang, X., Cheng, T., Wang, G. et al. Cloning and evolutionary analysis of tobacco MAPK gene family. Mol Biol Rep 40, 1407–1415 (2013). https://doi.org/10.1007/s11033-012-2184-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-2184-9