Abstract
The ATP-binding cassette (ABC) superfamily is a larger protein family with diverse physiological functions in all kingdoms of life. We identified 53 ABC transporters in the silkworm genome, and classified them into eight subfamilies (A–H). Comparative genome analysis revealed that the silkworm has an expanded ABCC subfamily with more members than Drosophila melanogaster, Caenorhabditis elegans, or Homo sapiens. Phylogenetic analysis showed that the ABCE and ABCF genes were highly conserved in the silkworm, indicating possible involvement in fundamental biological processes. Five multidrug resistance-related genes in the ABCB subfamily and two multidrug resistance-associated-related genes in the ABCC subfamily indicated involvement in biochemical defense. Genetic variation analysis revealed four ABC genes that might be evolving under positive selection. Moreover, the silkworm ABCC4 gene might be important for silkworm domestication. Microarray analysis showed that the silkworm ABC genes had distinct expression patterns in different tissues on day 3 of the fifth instar. These results might provide new insights for further functional studies on the ABC genes in the silkworm genome.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The ATP-binding cassette (ABC) transporters are one of the largest families of transmembrane proteins. They are found in all organisms from prokaryotes to eukaryotes. The core functional unit of ABC transporters consists of four domains: two cytoplasmic domains containing the highly conserved nucleotide-binding domain (NBD, also known as ABC) responsible for ATP-binding and hydrolysis, and two transmembrane domains (TMD) involved in the substrate translocation pathway [1]. The functional unit of prokaryotic ABC importers has a fifth domain, the substrate-binding protein (SBP) that contributes to the substrate specificity of ABC importers [2]. The NBD contains several highly conserved motifs: Walker A, Walker B and a third conserved sequence called the ABC signature, and the C motif, which is located between the two Walker motifs, just upstream of Walker B [3, 4]. The eukaryotic ABC genes are organized either as “full-size” transporters, symbolized as (TMD-NBD) 2, or as “half-size” transporters (TMD-NBD or NBD-TMD). Half transporters need to form homo- or hetero-dimers to compose a functional molecule [3, 5]. Based on gene sequence homology and NBD and TMD domain organization, eukaryotic ABC transporters can be divided into eight subfamilies (ABCA–ABCH), seven of which (ABCA–ABCG) exist in the human genome. The ABCH subfamily has been found in the Drosophila melanogaster genome for the first time [6].
Most of the known functions of ABC proteins are in membrane transport of substrates, including amino acids, sugars, lipids, ions, lipopolysaccharides, peptides, saccharides, metals, and chemotherapeutic drugs [7, 8]. The first eukaryotic ABC transporter identified was the multidrug resistance (MDR) efflux transporter responsible for MDR in cancer cells caused by decreased cellular drug accumulation [5, 9]. Subsequent studies revealed that ABC drug efflux transporters are widely present in most organisms. D. melanogaster mrp-1 has a novel gene structure containing two variable internal exons, resulting in 14 different multidrug resistance-associated protein (MRP) isoforms through variable exon substitution [10]. In Caenorhabditis elegans, four ABC proteins are involved in drug resistance: three P-glycoproteins (pgp-1, -3 and -4) and one MDR protein (mrp-1). Mutant strains of pgp-1, pgp-3 and mrp-1 are hypersensitive to the heavy metals cadmium and arsenite [11]. The expression of MRP transporters is usually associated with reduced efficacy of agricultural fungicides and herbicides [9, 12] and infection of pathogenic microorganisms [13].
In insects, ABC transporters are involved in transporting of pigments, and in development and drug resistance [14–16]. For example, the Drosophila White protein, dimerizes with the brown and scarlet to transport eye pigment precursors [14]. In Bombyx mori, Bmwh3 is equivalent to w-3, and is a homolog of the Drosophila white gene. The w-3 oe mutant silkworm has white eyes, white eggs and translucent larval skin [17]. The tobacco hornworm (Manduca sexta) can live entirely on tobacco. Previous studies reported that P-glycoprotein-like multidrug transporters play vital roles in excretion of dietary nicotine in the Malpighian (excretory) tubules of M. sexta, preventing influx of nicotine across the blood–brain barrier [18, 19].
Complete inventories of ABC proteins are available for many other organisms and an increasing number of ABC transporters have been characterized at the functional level. The silkworm is one of the most important genetic insect models, and has economic and scientific significance [20]. However, a genome-wide study on ABC genes has not yet been performed in silkworm. Such an integrated analysis of ABC genes could provide crucial clues for further studies on the putative physiological functions of ABC transporters in B. mori, such as the mechanisms of transport of toxic compounds and pigments. For this reason, we proposed a complete inventory of ABC transporters based on the available silkworm genome sequences [21]. Detailed sequence comparisons of each subfamily members with these of human, fruit fly and worm reveal their evolutionary relationships. We also studied microarray-based tissue expression profiles and analyzed genetic variations.
Materials and methods
Identification of ABC proteins in the silkworm
Silkworm ABC proteins of were identified using the model ABC-tran (accession PF00005) of the Pfam database and the HMMER program from the HMMER package [22, 23]. By searching a new assembly of the silkworm genome dataset with PF00005 of the PFAM HMM model, top hits for putative transporters were retained by high score and low E value (score ≥0 and E value ≤0.1). The most poorly matched (score ≤0 and E value ≥0.1) were checked by BLASTP on the National Centre for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/), retaining only those with highly significant matches to annotated ABC proteins or those with clear ATPase domains. This process was repeated until no new ABC proteins were found. The number and the positions of TMD in each ABC transporter were searched using the TMHMM Server v. 2.0. (http://www.expasy.ch/).
Classification of silkworm ABC transporters and phylogenetic analysis
The amino acid sequences of ATPase domains were extracted by performing Pfam searches at the Sanger Institute Web site (www.sanger.ac.uk/Software/Pfam/). We adopted the Human Genome Organization (HUGO)-approved subfamily designations [24, 25]. ATPase domains were used for phylogenetic analysis along with those of human transporters. Multiple sequence alignments were performed using ClustalX [26]. Neighbor-joining phylogenetic trees were reconstructed using MEGA 4.0 [27], and bootstrapping was with 1,000 replicates. The sorting results were further confirmed by BLAST as above. Phylogenetic analysis was performed with per subfamily proteins of silkworm and each subfamily of ABC proteins of from human, fruit fly and worm.
Genetic variations analysis of the silkworm ABC genes
Using 40 genomes from geographically different domesticated and wild silkworms, we evaluated genetic variations of ABC genes during silkworm domestication [28]. Based on the known 16 million Single nucleotide polymorphisms (SNPs) and 354 domestication-related genes in the silkworm varieties, we surveyed SNPs and domestication of the silkworm ABC genes using the PAML package to evaluate the pattern of positive selection for each ABC gene [29].
Microarray analysis
A genome-wide microarray with 22,987 oligonucleotides was constructed and analyzed using an experimental process described previously [30]. We found that 52 ABC genes had probes on the microarray, and surveyed their expression profiles in testis, ovary, fat body, midgut, hemocyte, Malpighian tubules, head, anterior/middle silk (A/MSG) gland and posterior silk gland (PSG) from silkworm individuals on day 3 of the fifth instar.
Results
Identification and characterization of ABC transporters in the silkworm genome
We identified 53 ABC genes in the silkworm genome (Table 1). These include the five previously identified ABC genes Bmwh3 (GenBank accession no. BAH03523), BmRLI (AB164193), subfamily F member 2 (NP_001040334), two partial sequences of wh1 (AAF61570) and wh2 (AAF61571) (Supplementary Table 1). The numbers of ABC genes were compared among B. mori, D. melanogaster, C. elegans and Homo sapiens. As shown in Table 1, in C. elegans, 60 ABC genes form the largest ABC data set. In vertebrates, a remarkable decrease in the number of ABC genes was observed: for instance, only 49 ABC genes have been identified in humans. In insects, the silkworm and D. melanogaster have similar number of ABC genes; however, the percentages of ABC genes are much higher than in C. elegans or H. sapiens.
Based on a high-quality genome map, 52 ABC genes were located on 18 different chromosomes. Interestingly, more than 60% of the silk worm ABC genes were clustered (Fig. 1). Only seven genes were found to be the only ABC gene on a chromosome. Most clusters contained members of the same subfamily: for instance, the ABCA genes on chromosomes 14 and 17, ABCB genes on chromosomes 1, the ABCC genes on chromosomes 12 and 15, and the ABCG genes on chromosomes 5, 10 and 12. The largest ABC cluster was found on chromosome 15, containing eight ABC genes with gaps of 5–3,000 kb between each other in a big scaffold. Approximate 20% of the silkworm ABC genes are on Chromosome 15; interestingly, eight belong to the ABCC subfamily. The silkworm ABC genes are suggested to have expanded by gene duplication. As previously reported, the ABCG6 gene, a homolog of the fruit fly white gene (known as Bmwh3), is located on Chromosome 10. BmABCC14 is located on scaffold1071, but cannot be mapped to any chromosome in the current assembly version.
Domain-based analysis and classification of the silkworm ABC transporters
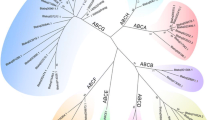
To analyze the structure of silkworm ABC transporters, the positions of the NBDs and TMDs were identified for predicted proteins. The silkworm ABC transporters were organized either as full transporters containing two TMDs and two NBDs or as half transporters containing any one of them (Fig. 2). Moreover, some ABC transporters contained only a single ATPase domain without a TMD, or two directly linked ATPase domains without a TMD. Based on the HUGO-approved subfamily designations, we performed phylogenetic analysis on the ATPase domains, and found that all 53 ABC proteins were grouped into eight subfamilies (A–H) (Supplementary Table 1), with full transporters were distributed among the ABCA, ABCB and ABCC families. And the half transporters can either occur in the TMD-NBD organization, as in the ABCA and ABCB families, or an NBD-TMD organization, as in the ABCG and ABCH subfamilies. In addition, the ABCE and ABCF subfamilies were entirely composed of proteins with two ATPase domains and no TM domains (Supplementary Table 1).
Phylogenetic analysis of silkworm ABC subfamilies
ABCA subfamily
The ABCA subfamily proteins were well-characterized full transporters. The silkworm ABCA subfamily contains nine members: one full transporter, four half-size members and four ABC proteins containing only an ATPase domain (Supplementary Table 1), suggesting that the ABCA subfamily has underwent rapid divergence. The evolutionary relationships of these proteins and H. sapiens, C. elegans and D. melanogaster ABCA transporters were shown in Supplementary Fig. 1. The BmABCA4 groups were in single large clade with HsABCA5, 6, 8, 9, 10 and a group of silkworm ABCA transporters (BmABCA1, 2 and 3), indicating that the four silkworm ABC genes are closely related to the HsABCA8 clusters on Chromosome 17q24 [6]. To date, HsABCA6, 8 and 10 have been considered to be involved in MDR, lipophilic drug transport and to be cholesterol-responsive genes [31]. Additionally, BmABCA5 clusters with Drosophila CG1819, BmABCA6 is a co-ortholog of Drosophila CG6052 and CG1718, and BmABCA8 groups together with C. elegans Abt-1 (Supplementary Fig. 1).
ABCB subfamily
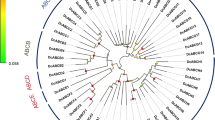
The ABCB subfamily includes two types of proteins: full transporters and half transporters. In the silkworm, the ABCB subfamily consists of five full transporters and four half transporters (Supplementary Table 1). As expected, phylogenetic analysis of ABCB subfamily showed that the full transporters were clustered into a group at the top of the tree, with two clades comprised of half transporters at the bottom (Fig. 3). All five full ABCB transporters appeared to be most closely related to the Drosophila biochemical defense genes (Dmr49, Dmr50 and Dmr65), and all these genes formed a larger cluster with the human Pgp (MDR/PGP) subfamily of ABC transporters, which have a wide range of functions including bile salt excretion from the liver and export of hydrophobics and steroids. As seen in from the phylogenetic tree, each of the four half transporters had one orthologous relationship with human mitochondrial transporters, which are involved in iron and Fe/S-cluster transport. The relationships are BmABCB6/HsABCB6, BmABCB7/HsABCB7 and BmABCB8/HsABCB8. BmABCB9 groups together with HsABCB10 with comparatively low bootstrap support (Fig. 3). HsABCB10 and HsABCB8 form a heterodimeric full transporter in the mitochondria [32]. HsABCB2/TAP1 and HsABCB3/TAP2 form a heterodimer to transport peptides into the endoplasmic reticulum (ER) lumen for peptide processing, resulting in presentation on the cell surface [33], However, we could not find orthologous relationships for any of them in the silkworm.
Phylogenetic tree of ABCB proteins in three eukaryotic genomes. Amino acid sequences were aligned using ClustalX and subjected to phylogenetic analysis using the neighor-joining method. Reliabilities of each branch point were assessed by the analysis of 1,000 bootstrap replicates. Bm Bombyx mori, Dm Drosophila melanogaster, Ce Caenorhabditis elegans, Hs Homo sapiens
ABCC subfamily
The C subfamily proteins are always full transporters. The diverse functions of human ABCC transporters include ion transport, toxin excretion, and cell surface receptors [6, 31]. In the silkworm genome, ABCC proteins form the largest subfamily, with 15 members with diverse structures (Supplementary Table 1). Phylogenetic analysis showed that BmABCC6 grouped with Drosophila CG5789 and clustered with the silkworm ABCC transporters (BmABCC1, 4 and 5), with BmABCC7 and BmABCC8 as neighboring genes (Supplementary Fig. 2). All six genes were located on chromosome 15, suggesting that the silkworm ABCC subfamily might have arisen from gene duplication. In addition, phylogenetic analysis indicated that BmABCC2 and BmABCC3 were closely related to HsABCC1 (Supplementary Fig. 2), HsABCC3 and Drosophila CG6214, an ortholog of human multidrug resistance-associated proteins MRP1, MRP2 and MRP3 [16]. BmABCC10 is a homolog of human ABCC10/MRP7. BmABCC9 is a homolog of human ABCC7 (CFTR), which functions as a chloride channel that is mutated in cystic fibrosis [34]. Interestingly, five neighboring ABCC genes (BmABCC11-15) were specific to silkworm and not homologous to other genes in other organisms (Supplementary Fig. 2).
ABCD, ABCE and ABCF subfamilies
The ABCD, ABCE and ABCF subfamilies in the silkworm contained a small number of ABC members with clear orthologous relationships. The ABCD subfamily contained two ABC proteins, which were half transporters with orientation TMD-NBD (Fig. 2). These ABC proteins are located in the peroxisome, where they are involved in fatty acid transport [35]. Phylogenetic analysis revealed that BmABCD1 clustered with DmCG2316. These are homologs of HsABCD1 and HsABCD2 with relatively low bootstrap support. BmABCD2 is a putative homolog of HsABCD3 (65% amino acid identity) (Supplementary Fig. 3), which is mutated in Zellweger syndrome 2 [36].
The proteins of subfamilies E and F are characterized by two ATPase domains and no TMD, so they are unlikely to function as transmembrane transporters. Only one gene (BmABCE1/BmRLI) was found on Chromosome 7 in the silkworm genome (Supplementary Table 1). Phylogenetic analysis showed that BmABCE1 is ortholog of HsABCE1(Supplementary Fig. 3), which promotes interferon activity [31]. We also found three ABCF transporters, BmABCF1, 2 and 3. Each had two ATPase domains and no TM domain (Supplementary Table 1). By phylogenetic analysis of ABCF transporters, BmABCF1 and BmABCF3 were orthologous with human HsABCF1 and HsABCF3 (61 and 63% amino acid identity). In addition, BmABCF2 was a homolog of HsABCF2 and Drosophila CG9281 (Supplementary Fig. 3), but its function is unclear.
The ABCG and ABCH subfamilies
The silkworm ABCG proteins are second largest subfamily, with 12 members, most of which were half ABC transporters with a reverse domain organization (NBD-TMD). Two ABCG proteins (BmABCG5 and BmABCG8) had only a single ATPase domain. Phylogenetic analysis revealed that BmABCG1-6 had clear orthologous relationships with Drosophila, while BmABCG7-12 were appeared to be silkworm-specific, and clustered in a special clade (Supplementary Fig. 4). BmABCG4 grouped with four Drosophila transporters, while BmABCG1 grouped with HsABCG1, HsABCG4, and two Drosophila transporters, Atet and CG3164. Drosophila Atet is reported to be expressed in trachea, where it is involved in transporting a small molecule after dimerization with a partner protein [37]. Additionally, BmABCG5 is a putative homolog of DmBrown (33% amino acid identity). BmABCG6 (also known as Bmwh3), a homolog of the Drosophila white gene, is involved in the transport of eye pigment precursors [38]. The silkworm ABCH subfamily had two half ABC transporters with a reverse domain organization similar to the ABCG subfamily (Fig. 2; Supplementary Table 1). Phylogenetic analysis revealed that two members clustered with Drosophila ABCH proteins (Supplementary Fig. 5). Furthermore, BmABCH1 was orthologous to Drosophila protein CG33970.
Genetic variations of silkworm ABC genes
In addition to phylogenetic analysis, we also surveyed variations in ABC genes during silkworm domestication. BmABCC4 is the only one of found in the 354 candidate domestication-related genes that are thought to be important during silkworm domestication [28]. We investigated the SNPs in coding sequences of ABC genes between domesticated and wild silkworm. In general, the number of SNPs per ABC gene was similar in the domesticated and wild silkworm groups (data not shown). We estimated the positive selection patterns of ABC genes during silkworm domestication using PAML software. As shown in Table 2, BmABCA6, BmABCB2, BmABCC6 and BmABCG1 were significantly determined to be evolving under positive selection (P values for BmABCA6 = 0.026881; BmABCB2 = 0.000108599; BmABCC6 = 0.006562637; BmABCG1 = 0.003428).
Expression profile of ABC genes
The expression patterns of ABC genes were surveyed using microarrays constructed with 22,987 oligonucleotides as 70-mer probes covering present and predicted genes in the silkworm genome [30]. The expression patterns of 52 ABC genes were analyzed in 10 tissues on day 3 of fifth instar larvae. We detected 28 genes, covering the eight subfamilies from ABCA to H, and found all were expressed in at least one tissue (Fig. 4). The silkworm ABC genes had distinct expression patterns, according to the microarray data. For example, five genes were widely expressed in every tissue, most notably BmABCE1 (BmRLI). Only one member of the ABCE subfamily was consistently highly expressed in all surveyed samples. In addition, both BmABCF3 and BmABCF1 were detected in the surveyed tissues. Seven ABC genes were considered to be tissue-specific: for example, BmABCB3, BmABCG5, and BmABCG9 were specifically expressed in Malpighian tubules (Fig. 4), which are insect excretory and osmoregulatory organs. Three genes were expressed specifically in testis (BmABCA8, BmABCC8 and BmABCC14). BmABCA2 was preferentially expressed in the PSG, suggesting that these transporters may be involved in silk formation.
Discussion
ABC transporters are one of the largest families of transmembrane proteins and are involved in the fundamental membrane transport of a variety of substrates. To date, they have been identified in many organisms based on genome sequencing projects. In this study, we have identified 53 ABC genes, representing approximately 0.36% of the total genes in a new assembly of B. mori genome. The silkworm contains representative members of each subfamily (A–H). As in the human and fruit fly genomes [6], the silkworm ABC genes are widely dispersed in the genome. Analysis of chromosomal locations showed that most of the gene clusters were composed of genes from the same subfamily and were arranged in a head-to-tail fashion, for example, the ABCC and ABCG subfamilies, which represent the largest and second largest silkworm ABC subfamilies (Table 1). The ABCC genes were mostly located on chromosomes 15 and 12, while the ABCG genes were clustered on chromosomes 10 and 12 (Fig. 1). These results revealed that the ABCC and ABCG subfamilies likely arose from gene duplication, a finding similar to Donly [39]. The ABCC and ABCG subfamilies have two groups of silkworm-specific genes, many located on the same chromosome. One example is BmABCG7 and BmABCG11, and BmABCG10 and BmABCG8, which occur as tandem repeats on chromosome 12. This indicated that tandem repeat distribution is a possible factor in the expansion of silkworm ABC genes. Therefore, genes originating from gene duplication might have adapted during evolution and have specific biological roles in the silkworm. However, this hypothesis requires further validation.
The silkworm ABCA subfamily appears to have undergone rapid divergence. A classic example of this is the animal ABCA subfamilies, which are composed entirely of full transporters (12 full transporters in humans), while the silkworm ABCA subfamily contains only one full transporter. The silkworm ABC genes might have suffered a massive loss of the last copies of half transporters during evolution. This trend has been seen in Dictyostelium discoideum and plants [40]. By contrast, the ABCD, ABCE and ABCF subfamilies seem to be more conserved in the silkworm. Most of these genes are also conserved in humans, flies and worms [24]. The human ABCD genes (HsABCD1/ALDP and HsABCD3) play critical roles in importing fatty aids and/or fatty acyl-coenzyme into the peroxisome. Mutations in HsABCD1 cause adrenoleukodystrophy (ALD), an x-linked metabolic disorder characterized by neurodegeneration and adrenal deficiency [41]. HsABCD3 is also a member of the ALD subfamily. Given the orthologous relationship with HsABCD1 and HsABCD2, BmABCD1 and BmABCD2 might be members of ALD involved in fatty acid transport. All the organisms have one ABCE and three ABCF genes. Each of the silkworm ABCE and ABCF genes has a clear ortholog in the human genome and other species (Supplementary Fig. 3). The human ABCE1 protein has been identified as a ribonuclease L inhibitor (RLI), a regulatory protein whose co-expression inhibits the binding of 2-5A by the endogenous RNase L [42]. BmABCE1 (BmRIL) has been cloned as a homolog of RLI, but its function has not been determined. The well-known ABCF protein is Saccharomyces cerevisiae GCN20, which mediates the activation of the eIF-2 alpha kinase. The human ABCF1 (GCN20 homolog) appears to have a similar role [24]. BmABCF1 is closely related to HsABCF1 and is a candidate for a translational regulator. Microarray-based expression analysis revealed that all ABCE and ABCF genes were highly expressed in surveyed samples on day 3 of fifth instar larvae, a key stage for silk protein synthesis and preparation for metamorphosis. This suggested that these highly conserved genes may have fundamental roles in the silkworm. This coincides with the evolutionarily conserved features of these ABC transporters in the silkworm.
The ABCB subfamily is best known to be associated with an MDR phenotype in cancer cells. Our analysis revealed five ABCB genes (BmABCB1-5) that appear to be closely related to fly MDR genes. In insects, the Drosophila genes Dmr49, Dmr50 and Dmr65 are homologs of mammalian MDR genes [43, 44]. Dmr49 is involved in biochemical defense against toxins, while the Dmr65 is considered to be related to α-amanitin resistance in Drosophila [44, 45]. Therefore, we hypothesized that these MDR-related genes might be involved in biochemical defense. Given that the five ABCB genes of silkworm are most closely related to the human and fly MDR genes, assigning them to the MDR subfamily of the ABC proteins according to their putative physiological roles seems reasonable. Additionally, microarray experiments indicated that many ABC genes were tissue-specific, exhibiting a strong relevance to the physiological functions of the corresponding tissues. For example, BmABCB3 was specifically highly expressed in Malpighian tubules (Fig. 4), which are the excretory and osmoregulatory organs of insects. With our phylogentic analysis, BmABCB3 might be involved in detoxification in the silkworm Malpighian tubules. Moreover, BmABCB5 was especially highly expressed in testis. Previously more than 1,000 genes were reported to be expressed specifically in the testis and their function might be in spermatogenesis [30]. This is suggested that these ABC transporters may be related to silkworm sex development of the silkworm.
ABCC transporters have diversified biological functions, including ion transport, toxin secretion and acting as cell surface receptors [46]. The largest group of ABC proteins is MRPs, most of which are involved in export of toxic compounds. Phylogenetic analysis revealed that BmABCC2 and BmABCC3 are closely related to the human MRPs (MRP1, MRP2 and MRP3) (Supplementary Fig. 2). Recently, these transporters have been shown to be important in immune processes, by translating several small proinflammatory molecules. For example, prostaglandins (PGs) are exported by human MRP4 [47, 48]. LTC4 is involved in the proinflammatory effect on inflammatory cells and is transported by MRP1 [49], MRP2 [50], MRP3 [51] or MRP4 [52]. Glutathione (GSH) is an important cellular antioxidant in cellular detoxification pathways, and its level is mediated by MRP1 in tumor cells [53]. BmABCC10 is a homolog of HsABCC10/MRP7, however, both contain an additional N-terminal series of five transmembrane spans called TMD0, whereas the TMD0 (TMD-NBD)2 arrangement is present only in the ABCC family. Cystic fibrosis transmembrane conductance regulator (CFTR) and sulfonyl urea receptor (SUR) are two other ABCC subfamilies found in eukaryotes that function as ion channels. In B. mori, BmABCC9 is a candidate for the CFTR gene, and this might also be the case in silkworm. However, the silkworm genome lacked homologs to the SUR genes. The expression patterns of the silkworm ABCC genes were also distinct. For example, BmABCC14, a silkworm-specific gene, was specially expressed in testis, as was BmABCC8. However, BmABCC10 was expressed in every investigated sample; furthermore, several ABCC genes were differently expressed in different tissues. The expansion of ABCC genes and their diversification in expression pattern might be important in silkworm evolution.
Although some ABCG transporters such as the human ABCG2 also confer an MDR phenotype in some cancer [24, 54], no orthologous genes were found in the silkworm. The most intensive studies on insect ABCG proteins is in the Drosophila white–brown complex (white, scarlet and brown), which is reported to be critical for transporting eye pigment precursors [6, 55]. In the silkworm, knocking down Bmwh3 (BmABCG6) by RNAi led to translucent larval skin and white eggs [56]. A recent study revealed that Bmwh3 has a single-base deletion in exon 2 and a premature stop codon at the 5′ end of exon 3 in the w-3 oe mutant, which leads to white eyes, white eggs and translucent larval skin. Moreover, Bmwh3 is responsible for transport of ommochrome precursors and uric acid into pigment granules and urate granules [17]. In addition, the Drosophila brown and scarlet genes are required subunits for deposition of pteridine pigments [14]. The phylogenetic tree illustrates that BmABCG5 is an ortholog of Drosophila brown genes. It was specifically highly expressed in the Malpighian tubules (Fig. 4). However, the silkworm lacked a clearly orthologous relationship to the Drosophila scarlet gene. The ABCH subfamily has been identified in Drosophila for the first time [24]. However, the function of ABCH transporters is unclear. Therefore, no detailed function predictions can be made about the silkworm ABCH genes based on phylogeny alone.
The silkworm has been domesticated for more than 5,000 years, and has evolved complete dependence on humans for survival through human selection [28]. The complete re-sequencing of 40 Bombyx genomes has identified 354 candidates for genes that might have been important for silkworm domestication [28]. Interestingly, ABCC4 is a domestication-related gene, and was highly expressed in the midgut (Fig. 4). The silkworm midgut is the location of nutrient digestion and absorption and represents the first line of resistance and immune response. Consequently, we consider that BmABCC4 might have been important in the midgut during silkworm domestication. In addition, positive selection of duplicated genes was found in the ABCB subfamily, which is best known as associated with acquiring an MDR phenotype. Moreover, genetic variation analysis revealed that four ABC genes (BmABCA6, BmABCB2, BmABCC6 and BmABCG1) were evolving under positive selection (Table 2), suggesting functional diversification of ABC adaptive evolution in silkworm domestication.
In conclusion, we carried out a comprehensive analysis of protein structure and evolution of 53 ABC transporters in the silkworm, and surveyed their expression patters by microarray. The ABCC subfamily of the silkworm has expanded more than in other species. The presence of genes similar to MDR and human MRP genes has deep implications for biochemical defense. Members of the ABCE and ABCF subfamilies were evolutionarily highly conserved, and expressed in all the surveyed samples, suggesting that these ABC proteins have essential roles in silkworm. The domestication-related ABC genes likely were involved in silkworm domestication. These data provide an overview of silkworm ABC transporters, which will facilitate further research on these genes.
References
Schneider E, Hunke S (1998) ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol Rev 22(1):1–20
Berntsson RP, Smits SH, Schmitt L, Slotboom DJ, Poolman B (2010) A structural classification of substrate-binding proteins. FEBS Lett 584(12):2606–2617
Hyde SC, Emsley P, Hartshorn MJ, Mimmack MM, Gileadi U, Pearce SR, Gallagher MP, Gill DR, Hubbard RE, Higgins CF (1990) Structural model of ATP-binding proteins associated with cystic fibrosis, multidrug resistance and bacterial transport. Nature 346(6282):362–365
Walker JE, Saraste M, Runswick MJ, Gay NJ (1982) Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J 1(8):945–951
Dean M (2009) ABC transporters, drug resistance, and cancer stem cells. J Mammary Gland Biol Neoplasia 14(1):3–9
Dean M, Rzhetsky A, Allikmets R (2001) The human ATP-binding cassette (ABC) transporter superfamily. Genome Res 11(7):1156–1166
Higgins CF (1992) ABC transporters: from microorganisms to man. Annu Rev Cell Biol 8:67–113
Klein I, Sarkadi B, Varadi A (1999) An inventory of the human ABC proteins. Biochim Biophys Acta 1461(2):237–262
Gottesman MM, Fojo T, Bates SE (2002) Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer 2(1):48–58
Grailles M, Brey PT, Roth CW (2003) The Drosophila melanogaster multidrug-resistance protein 1 (MRP1) homolog has a novel gene structure containing two variable internal exons. Gene 307:41–50
Broeks A, Gerrard B, Allikmets R, Dean M, Plasterk RH (1996) Homologues of the human multidrug resistance genes MRP and MDR contribute to heavy metal resistance in the soil nematode Caenorhabditis elegans. EMBO J 15(22):6132–6143
de Waard MA, Andrade AC, Hayashi K, Schoonbeek HJ, Stergiopoulos I, Zwiers LH (2006) Impact of fungal drug transporters on fungicide sensitivity, multidrug resistance and virulence. Pest Manag Sci 62(3):195–207
Webber MA, Piddock LJ (2003) The importance of efflux pumps in bacterial antibiotic resistance. J Antimicrob Chemother 51(1):9–11
Mackenzie SM, Brooker MR, Gill TR, Cox GB, Howells AJ, Ewart GD (1999) Mutations in the white gene of Drosophila melanogaster affecting ABC transporters that determine eye colouration. Biochim Biophys Acta 1419(2):173–185
Ricardo S, Lehmann R (2009) An ABC transporter controls export of a Drosophila germ cell attractant. Science 323(5916):943–946
Tarnay JN, Szeri F, Ilias A, Annilo T, Sung C, Le Saux O, Varadi A, Dean M, Boyd CD, Robinow S (2004) The dMRP/CG6214 gene of Drosophila is evolutionarily and functionally related to the human multidrug resistance-associated protein family. Insect Mol Biol 13(5):539–548
Komoto N, Quan GX, Sezutsu H, Tamura T (2009) A single-base deletion in an ABC transporter gene causes white eyes, white eggs, and translucent larval skin in the silkworm w-3(oe) mutant. Insect Biochem Mol Biol 39(2):152–156
Murray CL, Quaglia M, Arnason JT, Morris CE (1994) A putative nicotine pump at the metabolic blood–brain barrier of the tobacco hornworm. J Neurobiol 25(1):23–34
Gaertner LS, Murray CL, Morris CE (1998) Transepithelial transport of nicotine and vinblastine in isolated malpighian tubules of the tobacco hornworm (Manduca sexta) suggests a P-glycoprotein-like mechanism. J Exp Biol 201(18):2637–2645
Zhou Z, Yang H, Zhong B (2008) From genome to proteome: great progress in the domesticated silkworm (Bombyx mori L.). Acta Biochim Biophys Sin (Shanghai) 40(7):601–611
Xia Q, Zhou Z, Lu C, Cheng D, Dai F, Li B, Zhao P, Zha X, Cheng T, Chai C, Pan G, Xu J, Liu C, Lin Y, Qian J, Hou Y, Wu Z, Li G, Pan M, Li C, Shen Y, Lan X, Yuan L, Li T, Xu H, Yang G, Wan Y, Zhu Y, Yu M, Shen W, Wu D, Xiang Z, Yu J, Wang J, Li R, Shi J, Li H, Su J, Wang X, Zhang Z, Wu Q, Li J, Zhang Q, Wei N, Sun H, Dong L, Liu D, Zhao S, Zhao X, Meng Q, Lan F, Huang X, Li Y, Fang L, Li D, Sun Y, Yang Z, Huang Y, Xi Y, Qi Q, He D, Huang H, Zhang X, Wang Z, Li W, Cao Y, Yu Y, Yu H, Ye J, Chen H, Zhou Y, Liu B, Ji H, Li S, Ni P, Zhang J, Zhang Y, Zheng H, Mao B, Wang W, Ye C, Wong GK, Yang H (2004) A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science 306(5703):1937–1940
Bateman A, Birney E, Durbin R, Eddy SR, Finn RD, Sonnhammer EL (1999) Pfam 3.1: 1313 multiple alignments and profile HMMs match the majority of proteins. Nucleic Acids Res 27(1):260–262
Eddy SR (1995) Multiple alignment using hidden Markov models. Proc Int Conf Intell Syst Mol Biol 3:114–120
Dean M, Annilo T (2005) Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu Rev Genomics Hum Genet 6:123–142
Verrier PJ, Bird D, Burla B, Dassa E, Forestier C, Geisler M, Klein M, Kolukisaoglu U, Lee Y, Martinoia E, Murphy A, Rea PA, Samuels L, Schulz B, Spalding EJ, Yazaki K, Theodoulou FL (2008) Plant ABC proteins—a unified nomenclature and updated inventory. Trends Plant Sci 13(4):151–159
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25(24):4876–4882
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (mega) software version 4.0. Mol Biol Evol 24(8):1596–1599
Xia Q, Guo Y, Zhang Z, Li D, Xuan Z, Li Z, Dai F, Li Y, Cheng D, Li R, Cheng T, Jiang T, Becquet C, Xu X, Liu C, Zha X, Fan W, Lin Y, Shen Y, Jiang L, Jensen J, Hellmann I, Tang S, Zhao P, Xu H, Yu C, Zhang G, Li J, Cao J, Liu S, He N, Zhou Y, Liu H, Zhao J, Ye C, Du Z, Pan G, Zhao A, Shao H, Zeng W, Wu P, Li C, Pan M, Yin X, Wang J, Zheng H, Wang W, Zhang X, Li S, Yang H, Lu C, Nielsen R, Zhou Z, Xiang Z (2009) Complete re-sequencing of 40 genomes reveals domestication events and genes in silkworm (Bombyx). Science 326(5951):433–436
Yang Z (1997) PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci 13(5):555–556
Xia Q, Cheng D, Duan J, Wang G, Cheng T, Zha X, Liu C, Zhao P, Dai F, Zhang Z, He N, Zhang L, Xiang Z (2007) Microarray-based gene expression profiles in multiple tissues of the domesticated silkworm Bombyx mori. Genome Biol 8(8):R162
Vasiliou V, Vasiliou K, Nebert DW (2009) Human ATP-binding cassette (ABC) transporter family. Hum Genomics 3(3):281–290
Hogue DL, Liu L, Ling V (1999) Identification and characterization of a mammalian mitochondrial ATP-binding cassette membrane protein. J Mol Biol 285(1):379–389
Abele R, Tampe R (2004) The ABCs of immunology: structure and function of TAP, the transporter associated with antigen processing. Physiology (Bethesda) 19:216–224
Rommens JM, Iannuzzi MC, Kerem B, Drumm ML, Melmer G, Dean M, Rozmahel R, Cole JL, Kennedy D, Hidaka N et al (1989) Identification of the cystic fibrosis gene: chromosome walking and jumping. Science 245(4922):1059–1065
Shani N, Valle D (1998) Peroxisomal ABC transporters. Methods Enzymol 292:753–776
Mosser J, Lutz Y, Stoeckel ME, Sarde CO, Kretz C, Douar AM, Lopez J, Aubourg P, Mandel JL (1994) The gene responsible for adrenoleukodystrophy encodes a peroxisomal membrane protein. Hum Mol Genet 3(2):265–271
Kuwana H, Shimizu-Nishikawa K, Iwahana H, Yamamoto D (1996) Molecular cloning and characterization of the ABC transporter expressed in Trachea (ATET) gene from Drosophila melanogaster. Biochim Biophys Acta 1309(1–2):47–52
Abraham EG, Sezutsu H, Kanda T, Sugasaki T, Shimada T, Tamura T (2000) Identification and characterisation of a silkworm ABC transporter gene homologous to Drosophila white. Mol Gen Genet 264(1–2):11–19
Labbe R, Caveney S, Donly C (2011) Genetic analysis of the xenobiotic resistance-associated ABC gene subfamilies of the Lepidoptera. Insect Mol Biol 20(2):243–256
Anjard C, Loomis WF (2002) Evolutionary analyses of ABC transporters of Dictyostelium discoideum. Eukaryot Cell 1(4):643–652
Berger J, Gartner J (2006) X-linked adrenoleukodystrophy: clinical, biochemical and pathogenetic aspects. Biochim Biophys Acta 1763(12):1721–1732
Zhou A, Hassel BA, Silverman RH (1993) Expression cloning of 2–5A-dependent RNAase: a uniquely regulated mediator of interferon action. Cell 72(5):753–765
Gerrard B, Stewart C, Dean M (1993) Analysis of Mdr50: a Drosophila P-glycoprotein/multidrug resistance gene homolog. Genomics 17(1):83–88
Wu CT, Budding M, Griffin MS, Croop JM (1991) Isolation and characterization of Drosophila multidrug resistance gene homologs. Mol Cell Biol 11(8):3940–3948
Begun DJ, Whitley P (2000) Genetics of alpha-amanitin resistance in a natural population of Drosophila melanogaster. Heredity 85(2):184–190
Borst P, Evers R, Kool M, Wijnholds J (2000) A family of drug transporters: the multidrug resistance-associated proteins. J Natl Cancer Inst 92(16):1295–1302
Reid G, Wielinga P, Zelcer N, van der Heijden I, Kuil A, de Haas M, Wijnholds J, Borst P (2003) The human multidrug resistance protein MRP4 functions as a prostaglandin efflux transporter and is inhibited by nonsteroidal antiinflammatory drugs. Proc Natl Acad Sci U S A 100(16):9244–9249
van de Ven R, Oerlemans R, van der Heijden JW, Scheffer GL, de Gruijl TD, Jansen G, Scheper RJ (2009) ABC drug transporters and immunity: novel therapeutic targets in autoimmunity and cancer. J Leukoc Biol 86(5):1075–1087
Leier I, Jedlitschky G, Buchholz U, Cole SP, Deeley RG, Keppler D (1994) The MRP gene encodes an ATP-dependent export pump for leukotriene C4 and structurally related conjugates. J Biol Chem 269(45):27807–27810
Cui Y, Konig J, Buchholz JK, Spring H, Leier I, Keppler D (1999) Drug resistance and ATP-dependent conjugate transport mediated by the apical multidrug resistance protein, MRP2, permanently expressed in human and canine cells. Mol Pharmacol 55(5):929–937
Zeng H, Liu G, Rea PA, Kruh GD (2000) Transport of amphipathic anions by human multidrug resistance protein 3. Cancer Res 60(17):4779–4784
Rius M, Hummel-Eisenbeiss J, Keppler D (2008) ATP-dependent transport of leukotrienes B4 and C4 by the multidrug resistance protein ABCC4 (MRP4). J Pharmacol Exp Ther 324(1):86–94
Laberge RM, Karwatsky J, Lincoln MC, Leimanis ML, Georges E (2007) Modulation of GSH levels in ABCC1 expressing tumor cells triggers apoptosis through oxidative stress. Biochem Pharmacol 73(11):1727–1737
Kusuhara H, Sugiyama Y (2007) ATP-binding cassette, subfamily G (ABCG family). Pflugers Arch 453(5):735–744
Chen H, Rossier C, Lalioti MD, Lynn A, Chakravarti A, Perrin G, Antonarakis SE (1996) Cloning of the cDNA for a human homologue of the Drosophila white gene and mapping to chromosome 21q22.3. Am J Hum Genet 59(1):66–75
Quan GX, Kanda T, Tamura T (2002) Induction of the white egg 3 mutant phenotype by injection of the double-stranded RNA of the silkworm white gene. Insect Mol Biol 11(3):217–222
Acknowledgments
This work was supported by grants from the National Basic Research Program of China (No. 2012CB114600), Fundamental Research Funds for the Central Universities (No. CDJZR10290003), the National Natural Science Foundation (No. 30901054 and 31001034), and the Chongqing Natural Science Foundation (No. 2009BB1368). We gratefully acknowledge the International Science Editing for editing the manuscript. We also gratefully thank editors and anonymous referees for their recommendations and useful comments.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xie, X., Cheng, T., Wang, G. et al. Genome-wide analysis of the ATP-binding cassette (ABC) transporter gene family in the silkworm, Bombyx mori . Mol Biol Rep 39, 7281–7291 (2012). https://doi.org/10.1007/s11033-012-1558-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-1558-3