Abstract
The complex microbiome of the rumen functions as an effective system for the conversion of plant cell wall biomass to microbial proteins, short chain fatty acids and gases. In this study, metagenomic approaches were used to study the microbial populations and metabolic potential of the microbial community. DNA was extracted from Surti Buffalo rumen samples (four treatments diet) and sequenced separately using a 454 GS FLX Titanium system. We used comparative metagenomics to examine metabolic potential and phylogenetic composition from pyrosequence data generated in four samples, considering phylogenetic composition and metabolic potentials in the rumen may remarkably be different with respect to nutrient utilization. Assignment of metagenomic sequences to SEED categories of the Metagenome Rapid Annotation using Subsystem Technology (MG-RAST) server revealed a genetic profile characteristic of fermentation of carbohydrates in a high roughage diet. The distribution of phylotypes and environmental gene tags (EGTs) detected within each rumen sample were dominated by Bacteroidetes/Chlorobi, Firmicutes and Proteobacteria in all the samples. The results of this study could help to determine the role of rumen microbes and their enzymes in plant polysaccharide breakdown is fundamental to understanding digestion and maximising productivity in ruminant animals.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Livestock production in India is subsidiary to plant agriculture. In tropical countries, ruminants are fed on lignocellulosic by-products like cereal straws, tree foliages and cakes of oil seeds. Ruminants digest such plant materials by microbial processes. The rumen is characterized by its high microbial population density, high diversity and complexity of interactions. Bacteria are predominate in the rumen, along with a variety of anaerobic protozoa, archaea and fungi [1] and the associated occurrence of bacteriophage is also well documented [2]. The use of small subunit (SSU) rRNA gene sequence analysis has allowed a more complete description of the rumen microbiome and these inventories have demonstrated that a large microbial component remains uncharacterized and uncultured [3], especially a high proportion of the fibrolytic bacteria [4]. The rumen habitat contains a consortium of microorganisms that harbour the complex lignocellulosic degradation system for the microbial attachment and digestion of plant biomass. However, the complex chemical processes required to break down the plant cell wall are rarely carried out by a single species. Evidence also suggests that the most important organisms and gene sets involved in the most efficient hydrolysis of plant cell wall are associated with the fiber portion of the rumen digesta [5].
The bovine rumen provides a unique genetic resource for the discovery of plant cell wall-degrading microbial enzymes for use in biofuel production, presumably because of co-evolution of microbes and plant cell wall types [6]. However, there are limitations to metagenome mining [7], and the number of clones needed to represent the entire metagenome is staggering [8]. Nonetheless, this approach does allow one to begin to harvest the remarkable and vast diversity present in a given metagenome [9].
The sequencing of genomes from several hundred bacterial and numerous eukaryotic species have laid the foundation for generating genomic sequence data from whole environments avoiding a culturing step. This approach, also known as ‘‘metagenomics’’, is defined as the genomic analysis by direct extraction and cloning of DNA from an assemblage of microorganisms [10]. One approach has been the use of the pyrosequence technology to increase the depth of SSU rRNA surveys by sequencing amplicons from the variable region of the SSU molecule [11–13]. The second approach uses random sample pyrosequencing to generate environmental gene tags (EGTs) and protein families [14] from microbiomes to highlight significant differences in metabolic potential in each environment [15]. Moreover, this massive depth metagenomic sequencing is an invaluable complement to what has already been learned about lignocellulose degradation in the rumen. With the help of this technology, buffalo rumen (Bubalus bubalis) can be studied for diet and management dependent changes through metagenomic investigations on the microbial community and metabolic potential. Data presented in this study is a comparative metagenome analysis of the Surti buffalo microbiome under different diet composition using the inexpensive, massively parallel, and rapid method of pyrosequencing.
Materials and methods
Rumen sampling
The permission of the Committee for the Purpose of Control and Supervision of Experiments on Animals (CPCSEA) was obtained prior to initiation of the study. Eight Surti buffaloes, 2–3 years of age and weighing approximately 200 kg, were divided into four groups of two animals and were housed at the Department of Animal Nutrition, College of Veterinary Science and Animal Husbandry, Anand. Each group was given different treatment in form of ad lib total mixed ration (TMR) namely T1, T2, T3 and T4. Treatments consisted of a roughage to concentrate (R:C) of 100:0, 75:25, 50:50, and 25:75, respectively in individual feeding stalls. Concentrates were high-quality, low-fiber feeds that contained a high concentration of digestible energy per unit weight and volume. Under this study, the concentrate diet consisted of 20.11% crude protein, 10.28% crude fibre, 3.80% ether extract, 52.43% nitrogen free extract, 13.38% ash, 3.38% silica, 1.00% phosphorus and 1.22% calcium. All Animals were given free access for 2 h morning and evening, during which they had free access to drinking water. The feeding experiment was conducted for a period of 30 days. Sample of the rumen liquor (500 ml) was collected separately from individual animal on 30th day of experiment after 2 h of morning feeding by a suction pump using a flexible stomach tube as described earlier by Khampa et al. [16]. About 100 ml rumen fluid was passed through four layers of cheese cloth to remove particulate matter. Remaining rumen fluid was stored at −80°C for further study. Total genomic DNA was extracted from four groups following a protocol similar to the extraction of high molecular weight DNA with little modification (enzyme–chemical–glass bead lysis method) Liles et al. [17] and Qiagen stool kit protocol. DNA purity and concentration was analyzed by spectrophotometric quantification and gel electrophoresis.
Pyrosequencing and sequence analysis
Four samples were subjected to pyrosequencing (454 Life Sciences technology) based high throughput sequencer (GS FLX, Roche) as manufacture’s protocol. In brief, the samples were nebulised to generate fragments of size 500–800 bp. The sequencing library was prepared by applying adapters (A: 5′-CCATCTCATCCCTGCGTGTCCCATCTGTTCCCTCCCTGTCTCAG-3′ and B:5′CCTATCCCCTGTGTGCCTTGCCTATCCCCTGTTGCGTGTCTCAG-3′) on both ends of the fragments as described by manufacturer. Emulsion PCR was carried out to clonally amplify fragments on sequencing beads, followed by its recovery and loading onto Pico titre plate along with enzyme beads. Pyrosequencing was carried out for 200 cycles with the flow of A, T, G and C nucleotides sequentially and image was captured. Captured images were processed by image processing software to get sequencing reads. The data were analyzed using the SEED annotation engine (http://seed.sdsu.edu/FIG/index.cgi) [18] under metagenome rapid annotation using subsystem technology (MG-RAST) server. The sequences were compared using the BLASTX algorithm with an expected cut off of 1 × 10−5 [19]. Ribosomal Database Project (RDP) was used for robust bacterial classification and the European Ribosomal RNA database was used to classify eukaryotic and archaeal sequences. The sequences used in this article are freely available from the SEED platform (MG-RAST). The IDs used in this study are: 4445089.3, 4445091.3, 4445093.3 and 4446901.3.
Results and discussion
The overall goal of this study was to obtain a detailed characterization of the rumen microbiome of Surti buffalo in four different feeding regimes, with respect to both phylotype (ribosomal DNA gene tags) and functional content (environmental gene tags; EGTs). Dissimilar relationships were observed for EGTs hits against the SEED for Bacterial, Archaeal and Eukaryia in all four groups due to unequal distribution of contigs numbers (experimental bias). The numbers of SSU rRNA hits in the Surti buffalo metagenomic libraries were very rare. It was 0.02% in T1, 0.04% in T2, 0.13% in T3 and 0.21% in T4 for bacteria, respectively (Table 1). Although few SSU rRNA hits has been also reported by Brulc et al. [20] in cattle rumen microbiome. Unfortunately, no SSU rRNA hits were observed in Archaea (European Ribosomal Database) and Eukarya (European Ribosomal Database) in our microbiomes. Although three sequences (reads) of T2 group (0.0295%) were hit with SSU rRNA database of other than bacteria, archaea and eukarya (Table 1). These reads may belong to the host animal or sampling bias.
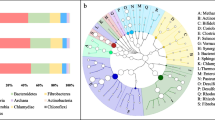
In theory, pyrosequencing should randomly sample the whole metagenome. From our analysis, there does not appear to be a bias with respect to location on the 16S rRNA molecule for the sequences retrieved from these four microbiome samples (Table 1). The phylogenetic composition of bacterial phyla for EGTs followed an unequal distribution for metagenomic libraries (Fig. 1), again highlighting the randomness of the sequenced libraries, as expected for a random sampling of genes. Additionally, our previous report based on Sanger sequencing [21] showed that phylogenetic composition of Surti buffalo rumen fed green fodder Napier bajra 21 (Pennisetum purpureum), mature pasture grass (Dicanthium annulatum), and concentrate mixture by partial 16S rRNA gene libraries were mostly belonged to Firmicutes, also known as the low G + C group and an unidentified group.
Phylogenic composition of bacterial phyla from four pyrosequenced environmental gene tags (EGTs). The percent of sequences in each of the bacterial phyla from A (T1, 100% roughage), B (T2, 75% roughage), C (T3, 50% roughage) and D (T4, 25% roughage) microbiomes are shown. The BLASTX cutoff of EGTs is 1 × 10−5 with a minimum length of 50 bp
Further insights into the diversity within the four different treatment groups of buffalo rumen metagenomic samples were obtained by comparing the number of EGTs (E value <1 × 10−5) of different bacterial phyla (Fig. 1). Sequence length is one of the primary factors in assessing similarity between sequences, and BLAST E values are dependent on both the length of the query sequence and the length of the database to which they are being compared [22]. However, this will affect the number of significant sequences found in the searches by a factor of two or more [23]. Pyrosequencing yielded more sequence than comparable Sanger sequencing, more than compensating for these missing sequences. The sequences missed in our searches are expected to be randomly distributed, and therefore, we are not expected to skew the comparative analysis. Indeed, while classifying EGTs from short pyrosequencing reads has been challenging, a report demonstrates that EGTs as short as 27 amino acids can accurately be classified with an average specificity ranging from 97% for Superkingdom to 93% for Order [24].
Bacterial specific EGTs represented approximately 70.73% in T1, 83.68% in T2, 79.24% in T3 and 64.88% in T4 of the total EGTs (Table 1) and the distribution of phylotypes fell predominantly into the bacteroidetes/Chlorobi, firmicutes and proteobacteria groups, regardless of the microbiome analyzed (Fig. 1). The distribution of EGTs from the bacteria is congruent with the distribution of SSU rRNA phylotypes, as was found with the Soudan Mine, cattle rumen and chicken ceacum microbiome studies [19, 20, 25]. Similar observations were also found in our previous study in chicken gut microbiome and goat rumen microbiome (data unpublished) by high throughput sequencing. Archaean EGTs constituted approximately 0.5%, 1.69%, 1.34% and 1.48% in T1, T2, T3, and T4 of the total EGTs, respectively (Table 1), matching well with previous estimates of Archaea numbers in the adult chicken cecum microbiome [26] and cattle rumen [20] microbiomes. The majority of Archaeal EGTs corresponded to Euryarchaeota with largest proportion corresponding to the methanogenic classes. Eukaryotic EGTs were 17.02%, 4.93%, 10.25% and 17.19% of total EGTs in T1, T2, T3 and T4, respectively (Table 1). The majority of eukarya ETGs were fungi/metazoan group. These EGT proportions were expected from our current knowledge of the rumen microbiome community structure. Interestingly, few other EGTs were also recovered in present microbiome in all four groups (Table 1).
Virus EGTs were very rare and primarily composed of double stranded (ds) DNA viruses in Surti buffalo rumen microbiome. The lack of viral sequences may be because of their extensive diversity and limited representation in public databases. Additionally, viral sequences may have been overlooked because they were not enriched in fluid during sampling procedures.
The subsystems-based annotations (SEED) database (MG-RAST) was utilized to gain a better understanding of these phylogenetic trends and to predict the metabolic potential (content of EGTs) of these microbiomes (Fig. 2). The EGTs proportions were similar to cattle rumen microbiome community structure [20]. The subsystems are annotated across genomes and are based on biochemical pathways, fragments of pathways, and clusters of genes that function together, or any group of genes considered to be related. Much of this analysis is dependent on sequence databases, and while we tried to avoid database bias by using multiple databases and alternative querying algorithms for analysis, we are aware that some sequences have no matched relatives in the databases, or are over-represented in the databases. Further, sequence identity does not always mean functional similarity and this may influence the interpretation of our results as minor sequence dissimilarities may represent functionally different or even a completely new functions.
SEED subsystem composition of four rumen microbiome A (T1; 100% roughage), B (T2; 75% roughage), C (T3; 50% roughage) and D (T4; 25% roughage) are shown. The percent of environmental gene tags (EGTs) of the SEED subsystems from the rumen microbiomes (T2) is shown. The BLASTX cut off for EGTs is 1 × 10−5
Figure 2 shows the metabolic profile (subsystems) of all four rumen microbioms (T1, T2, T3 and T4). The distribution of subsystems is strikingly different for the T1 (100% roughage) microbiome from the rumen of T4 (25% roughage), which is indicative of a community metabolism which has shifted away from a roughage digesta, more easily fermentable carbohydrate-based metabolism. It appears that animals from group T4 have low fiber metabolizing bacteria. This is consistent with the metabolism represented by the gammaproteobacteria. However, most of the gammaproteobacteria sequences were most similar to sequences from Psychrobacter-like organisms (from arctic samples), they probably are not from this genus, but rather from a close relative [27]. Our analysis were also similar with other microbiomes [25, 28], the Surti buffalo rumen microbiomes are dominated by carbohydrate metabolism in high fiber diet (100% roughage), lowest in high concentrate diet (75% concentrate), and are sparsely populated with genes for respiration, reflecting the more stable anoxic environment in the gastrointestinal tract (Fig. 3).
Carbohydrate metabolism subsystem composition of four rumen microbiomes in A (T1; 100% roughage), B (T2; 75% roughage), C (T3; 50% roughage) and D (T4; 25% roughage) are shown. The percent of environmental gene tags (EGTs) in each of the carbohydrate metabolism subsystems from the rumen microbiomes is shown. The BLASTX cut off for EGTs is 1 × 10−5
When looking solely at the Surti Buffalo (Bubuls bubalis) rumen microbiome and the SEED carbohydrate subsystem of all four groups, the central carbohydrate metabolism had a higher level in T1 group (26%) than in the T4 group (14%), while population size of monosaccharides EGTs were less 22% in T1 and higher 35% in T4. However, the carbohydrate EGTs of T2 group and T3 were similar proportion. This may be because of rumen pH together with microbial population, nature of substrates, environmental factors such as temperature, and the existence of cations and soluble carbohydrates have been suggested as factors governing bacterial attachment [29]. Ruminal pH is one of the most important of these factors, because the fibrolytic bacterial numbers are very sensitive to the pH change [30]. When ruminants are fed fiber- deficient rations, ruminal pH declines, microbial ecology is altered, and the animals become more susceptible to metabolic disorders [31].
While a limitation of the random sample pyrosequencing approach is the resulting short read lengths, we were able to assemble all of these reads into 280 total contigs of >500 nucleotides (39 from T1, 17 from T2, 187 from T3 and 37 from T4). Translations of these contigs (EGTs) were used for BLASTX analysis. The majority of these translations showed similarity with genes from the bacteroidetes, firmicutes and actinobacteria, the dominant taxa from these four microbiome. Similar observations have been also reported by Brulc et al. [20] in cattle rumen and Qu et al. [25] in chicken ceacum microbiome.
BlastX analysis indicates all contigs of four microbiome shared sequence similarities (27–100%) with different known organisms from the bacteroidetes, firmicutes, actinobacteria, proteobacteria and spirochetes etc., confirming the results from the non-assembled data and many contigs had sequence similarity with hypothetical proteins found in these four microbiome. However, four contigs (one from T1 and three from T3 group) were not showing any match with database, due to biasness in assembly process or may be novel.
Metagenomic analysis allows the relative abundances of all genes to be determined and used to generate a dataset for the assessment of the functional potential of each community [18, 32, 33]. Our exercise to assemble genes from four samples and most of the genes belonged to the Bacteroidetes/Chlorobi group suggests that this is an important phylum in Surti buffalo rumen, similar to that observed in studies of the human faecal microbiome [9, 34] cattle rumen [20] and chicken ceacum microbiome [25].
We conclude that the microbiome datasets presented herein represent the first assessment of the metabolic potential of the Surti buffalo (Bubulas bubalis) rumen microbiome at the functional gene level. As such, they represent a baseline for future studies and will be of great use in better understanding the large, complex, and dynamic microbial community of the rumen fluid, and the co-evolution/selection of microbes with their host and diet. It is clear that the composition and function the microbiome can be affected by various factors such as dietary ingredients, nutrient levels, environment, probiotic, and antibiotic treatments. Moreover, the gastrointestinal tract microbiome plays an important role in the growth and health of the host through its effects on gastrointestinal tract morphology, nutrition, pathogenesis of intestinal diseases, and immune responses. These microbiome data provide a critical genetic context for understanding animal nutrition, animal health and well-being. Additionally, the combined pyrosequence approach and subsystems-based annotations available in the SEED database allowed us to gain an understanding of the metabolic potential of these microbiomes. Sequence information was recovered in a comparative context based on the ecology of the microbial communities that inhabit the Surti rumen, which in the future will allow us to link metabolic potential to the identity of rumen microbes in their natural habitat.
References
Hespell RB, Akin DE, Dehority BA (1997). In: Mackie RI, White BA, Isaacson R (eds) Gastrointestinal microbiology, vol 2. Chapman and Hall, New York, pp 59–186
Klieve AV, Bauchop T (1988) Morphological diversity of ruminal bacteriophages from sheep and cattle. Appl Environ Microbiol 54:1637–1641
Tajima K, Aminov RI, Nagamine T, Matsui H, Nakamura M, Benno Y (2001) Diet-dependent shifts in the bacterial population of the rumen revealed with real-time PCR. Appl Environ Microbiol 67:2766–2774
Larue R, Yu Z, Parisi VA, Egan AR, Morrison M (2005) Novel microbial diversity adherent to plant biomass in the herbivore gastrointestinal tract, as revealed by ribosomal intergenic spacer analysis and rrs gene sequencing. Environ Microbiol 7:530–543
Forsberg CW, Cheng KJ, White BA (1997) Polysaccharide degradation in the rumen and large intestine. In: Mackie RI, White BA (eds) Gastrointestinal microbiology, vol 1. Chapman and Hall, New York, pp 319–379
Hess M, Sczyrba A, Egan R, Kim TW, Chokhawala H, Schroth G, Luo S, Clark DS, Chen, Zhang T, Mackie RI, Pennacchio LA, Tringe SG, Visel A, Woyke T, Wang Z, Rubin EM (2011) Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 331:463–467
Henne A, Daniel R, Schmitz RA, Gottschalk G (1999) Construction of environmental DNA libraries in Escherichia coli and screening for the presence of genes conferring utilization of 4-hydroxybutyrate. Appl Environ Microbiol 65:3901–3907
Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM (1998) Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol 5:245–249
Gill SR, Pop M, Deboy RT, Eckburg PB, Turnbaugh PJ, Samuel BS, Gordon JI, Relman DA, Fraser-Liggett CM, Nelson KE (2006) Metagenomic analysis of the human distal gut microbiome. Science 312:1355–1359
Handelsman J (2004) Metagenomics: application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev 68:669–685
Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta JM, Herndl GJ (2006) Microbial diversity in the deep sea and the underexplored “rare biosphere”. Proc Natl Acad Sci USA 103:12115–12120
Roesch LFW, Fulthorpe RR, Riva A, Casella G, Hadwin AKM, Kent AD, Daroub S, Camargo FAO, Farmerie WG, Triplett EW (2007) Pyrosequencing enumerates and contrasts soil microbial diversity. ISME J 1:283–290
Larsen N, Vogensen FK, Van Den Berg FWJ, Nielsen DS, Andreasen AS, Pedersen BK, Abu Al-Soud WMAF, Sorensen SJ, Hansen LH, Jakobsen M (2010) Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE 5:e9085
Tringe SG, von Mering C, Kobayashi A, Salamov AA, Chen K, Chang HW, Podar M, Short JM, Mathur EJ, Detter JC, Bork P, Hugenholtz P, Rubin EM (2005) Comparative metagenomics of microbial communities. Science 308:554–557
Edwards JE, McEwan NR, Travis AJ, Wallace RJ (2007) Characterization of the dynamics of initial bacterial colonization of nonconserved forage in the bovine rumen. FEMS Microbiol Ecol 62:323–335
Khampa S, Wanapat M, Wachirapakorn C, Nontaso N (2006) Effects of urea level and sodium di-malate in concentrate containing high cassava chip on ruminal fermentation efficiency, microbial protein synthesis in lactating dairy cows raised under tropical condition. Asian- Aust J Anim Sci 19:837–844
Liles MR, Williamson LL, Rodbumrer J, Torsvik V, Goodman RM, Handelsman J (2008) Recovery, purification, and cloning of highmolecular-weight DNA from soil microorganisms. Appl Environ Microb 74:3302–3305
Overbeek R, Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, Crécy-Lagard DV, Diaz N, Disz T, Edwards R et al (2005) The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 33:5691–5702
Edwards RA, Rodriguez-Brito B, Wegley L, Haynes M, Breitbart M, Peterson DM, Saar MO, Alexander S, Alexander EC, Rohwer F Jr (2006) Using pyrosequencing to shed light on deep mine microbial ecology. BMC Genomics 7:57–69
Brulc JM, Antonopoulos D, Berg-Miller ME, Wilson KM, Yannarell CA, Dinsdale AE, Edwards ER, Frank ED, Emerson BJ, Wacklin P, Coutinho MP, Henrissat B, Nelson EK, White AB (2009) Gene-centric metagenomics of the fiber-adherent bovine rumen microbiome reveals forage specific glycoside hydrolases. Proc Natl Acad Sci USA 106:1948–1953
Pandya PR, Singh KM, Parnerkar S, Tripathi AK, Mehta HH, Rank DN, Kothari RK, Joshi CG (2010) Bacterial diversity in the rumen of Indian Surti buffalo (Bubalus bubalis), assessed by 16S rDNA analysis. J Appl Genet 51:395–402
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Wommack KE, Bhavsar J, Ravel J (2008) Metagenomics: read length matters. Appl Environ Microbiol 74:1453–1463
Krause L, Diaz NN, Goesmann A, Kelley S, Nattkemper TW, Rohwer F, Edwards RA, Stoye J (2008) Phylogenetic classification of short environmental DNA fragments. Nucleic Acids Res. 36:2230–2239
Qu A, Brulc JM, Wilson MK, Law BF, Theoret JR, Joens LA, Konkel ME, Angly F, Dinsdale EA, Edwards RA et al (2008) Comparative metagenomics reveals host specific metavirulomes and horizontal gene transfer elements in the chicken cecum microbiome. PLoS ONE 3:e2945
Saengkerdsub S, Anderson RC, Wilkinson HH, Kim WK, Nisbet DJ, Ricke SC (2007) Identification and quantification of methanogenic archaea in adult chicken ceca. Appl Environ Microbiol. 73:353–356
Bozal N, Montes MJ, Tudela E, Guinea J (2003) Characterization of several Psychrobacter strains isolated from Antarctic environments and description of Psychrobacter luti sp nov and Psychrobacter fozii sp nov. Int J Syst Evol Microbiol 53:1093–1100
Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, Brulc JM, Furlan M, Desnues C, Haynes M, Krause L, Sala E, Sandin SA et al (2008) Functional metagenomic profiling of nine biomes. Nature 452:629–632
Miron J, Ben-Ghedalia D, Morrison M (2001) Invited review: adhesion mechanisms of rumen cellulolytic bacteria. J Dairy Sci 84:1294–1309
Sung HG, Kobayashi Y, Chang J, Ha A, Hwang H, Ha JK (2007) Low ruminal pH reduces dietary fiber digestion via reduced microbial attachment. Asian-Aust J Anim Sci. 20:200–207
Russell JB, Rychlik JL (2001) Factors that alter rumen microbial ecology. Science 292:1119–1122
Fierer N, Breitbart M, Nulton J, Salamon P, Lozupone C, Jones R, Robeson M, Edwards R, Felts B, Rayhawk S, Knight R, Rohwer F, Jackson RB (2007) Metagenomic and small-subunit rRNA analyses reveal the genetic diversity of bacteria, archaea, fungi, and viruses in soil. Appl Environ Microbiol 73:7059–7066
Wegley L, Edwards R, Rodriguez-Brito B, Liu H, Rohwer F (2007) Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ Microbiol 9:2707–2719
Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, Takami H, Morita H, Sharma VK, Srivastava TP, Taylor TD, Noguchi H, Mori H, Ogura Y, Ehrlich DS, Itoh K, Takagi T, Sakaki Y, Hayashi T, Hattori M (2007) Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 14:169–181
Acknowledgments
We thank Department of Biotechnology (DBT), Government of India and Rashtriya Krishi Vikas Yojana (RKVY) Government of Gujarat, for their financial support for this study.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Singh, K.M., Ahir, V.B., Tripathi, A.K. et al. Metagenomic analysis of Surti buffalo (Bubalus bubalis) rumen: a preliminary study. Mol Biol Rep 39, 4841–4848 (2012). https://doi.org/10.1007/s11033-011-1278-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-1278-0