Abstract
The gayal (Bos frontalis) is a rare semi-wild bovid species in which bovine spongiform encephalopathy (BSE) has not been reported. Polymorphisms of the prion protein gene (PRNP) have been correlated significantly with resistance to BSE. In this study, the coding region of PRNP was cloned and characterized in samples from 125 gayal. A total of ten single nucleotide polymorphisms (SNPs), including six silent mutations (C60T, G75A, A108T, G126A, C357T and C678T) and four mis-sense mutations (C8A, G145A, G461A and C756G), corresponding to amino acids T3K, G49S9, N154S and I252M were identified, revealing high genetic diversity. Three novel SNPs including C60T, G145A and C756G, which have not been reported previously in bovid species, were retrieved. There also was one insertion–deletion (187Del24) at the N-terminal octapeptide repeat region. Alignment of nucleotide and amino acid sequences showed a high degree of similarity with other bovid species. Using phylogenetic analyses it was revealed that gayal has a close genetic relationship with Zebu cattle. In short, preliminary information is provided about genotypes of the PRNP in gayal. This could assist with the study of the pathogenesis of transmissible spongiform encephalopathies and cross species transmission as well as a molecular breeding project for gayal in China.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Bovine spongiform encephalopathy (BSE) is one of the prion diseases, which also include scrapie in sheep, goats and mice, transmissible mink encephalopathy (TME) in mink, chronic wasting disease (CWD) in moose, deer and elk, feline spongiform encephalopathy (FSE) in cats, puma and cheetah, and also Creutzfeldt–Jacob disease (CJD), Gerstmann–Straussler–Sheinker (GSS) syndrome, fatal familial insomnia (FFI) and historical kuru in humans [1, 2]. A characteristic of BSE is the accumulation of a modified conformational neurotoxic isoform (PRNPSc) switching from the normal prion protein (PRNPC) in the bovine brain and spinal cord, forming deposits of β-amyloid resistant to proteolytic enzymes and resulting in the formation of specific histopathological lesions [3–5]. Normal prion protein is encoded by the prion protein gene (PRNP) which is a single gene with considerable genomic differences in human and domestic animal populations [6].
The gayal or mithun (Bos frontalis) is a rare semi-wild bovid species distributed throughout Bangladesh, Bhutan, China, India, Malaysia and Myanmar [7, 8]. In China, gayal is found predominantly in the narrow valleys of the Dulong and Nujiang Rivers and adjacent mountainous areas of Yunnan Province where they are described as ‘Dulong Cattle’ [9, 10]. Gayal has a chromosome complement of 2n = 58, which differs from those of cattle (Bos taurus, 2n = 60) and gaur (Bos gaurus, 2n = 56) [9, 11, 12]. Gayal browse tree leaves and graze grasses, bamboo leaves, reeds and other local plant species and reveals a very wide range of adaptive activities under the harsh conditions which range from cold to tropical belts [13, 14]. Due to the remoteness of their habitats and other ecological and sociopolitical factors, gayal remain one of the least studied ungulates [15] and has been catalogued in the red list of threatened species of the International Union for Conservation of Nature and Natural Resources (IUCN)—see www.redlist.org.
To date, the coding region of PRNP has been cloned and characterized extensively in European (B. taurus) [16–19] and Zebu (Bos indicus) cattle and their hybrids (B. indicus × B. taurus) [20], yak (Bos grunniens) [17], buffalo (Bubalus bubalis) [17], bison (Bison bison) and some wild ruminants [21–23]. However, for the “unique” gayal, the polymorphisms of PRNP have been investigated seldom. Although no natural case of BSE has been reported in gayal, it is considered to be very important to know more about the sequences of polymorphisms of PRNP in gayal. This could assist with the study of the pathogenesis of transmissible spongiform encephalopathies, cross species transmission and also a molecular breeding project for gayal in China. Therefore, in this report, we provide a detailed survey the PRNP coding region polymorphisms of gayal and have compared those with the sequences of PRNP from a range of bovid species.
Materials and methods
Sample collection
From 2005 to 2009, 125 blood or tissues samples were collected from apparently healthy gayal (B. frontalis) at the National Jiumudang Stud Gayal Farm (located at N25°47′02.6″, E099°05′56.5″ at an altitude of 2260 m above sea level), Dulong Town, Gongshan County, Yunnan Province, China. The appearance and mitochondrial DNA analyses of the animals were criteria used to exclude individuals showing evidence of interspecific hybridization with local Yunnan cattle (B. taurus) [24 and unpublished data]. All samples were snap frozen in liquid nitrogen then stored at −80°C pending analyses.
DNA extraction and polymerase chain reaction (PCR)
Genomic DNA was extracted from blood or tissues using standard proteinase K digestion followed by extraction with the phenol–chloroform method [25].
The PRNP sequence was amplified using previously described primers PRNP-CDS-F (5′-CTAGGGTCCCCACAAGAACAAG-3′) and PRNP-CDS-R (5′-ACGGGGCTGCAGGTAGATA-3′) [26]. The PCR was carried out in a reaction volume of 25 μl, containing 2.0 μl DNA (approximately 50 ng μl−1), 2.0 μl 250 μmol/l dNTPs, 2.5 μl buffer (MgCl2 in the buffer provided by the manufacture already), 2.0 μl 10 μmol/l forward primer, 2.0 μl 10 μmol/l reverse primer and 0.2 μl 10× Taq DNA polymerase (5 U/μl, Beijing TransGen Biotechnology Co., Ltd, China). Thermal cycling parameters were as follow: 5 min at 95°C, 35 cycles of amplification (45 s at 94°C, 45 s at 54°C, 60 s at 72°C), and finally 7 min at 72°C. The purified 125 PCR products were sequenced bidirectionally using an ABI3730 DNA Analyzer (Applied Biosystems Inc.) at the Sango Biotechnology Company (Shanghai, China).
Bioinformatic analyses
Genotype and allele frequencies were checked by counting directly, then the Hardy–Weinberg (H–W) equilibrium was assessed by application of the χ2 test. The haplotype frequencies were analyzed from the genotypic data by using the program package Haploview 4.0 [27]. Population genetic diversity indices, in terms of gene heterozygosity (He), gene homozygosity (Ho), effective allele numbers (Ne) and polymorphism information content (PIC) also were calculated using the previously described method by Nei and Li [28]. Nucleotide sequence alignment and construction of a phylogenetic tree were achieved by using the DNAStar program (Version 5.2.2) and MEGA software (Version 4.0). Protein motif prediction was made by using the PROSITE motif search tool http://www.expasy.org/prosite and http://www.cbs.dtu.dk/services/NetNGlyc/.
Results
Identification and distribution of the PRNP polymorphisms in the open reading frame (ORF)
The PCR amplicons covering the coding region of PRNP exon 3 were amplified successfully from 125 genomic DNAs and sequenced in both strands. The lengths of PCR amplicons were 1281 bp or 1257 bp. The sequences have been deposited in the GenBank database under accession numbers HQ262495 and HQ262496.
Ten single nucleotide polymorphisms (SNPs; C8A, C60T, G75A, A108T, G126A, G145A, C357T, G461A, C678T and C756G, numbered relative to the ORF) and one Indel polymorphism (187Del24) were found. Six SNPs (C60T, G75A, A108T, G126A, C357T and C678T) were silent mutations corresponding to encoding amino acids at D20, K25, T36, P42, N119 and I226, respectively. The remaining four SNPs (C8A, G145A, G461A and C756G) were mis-sense mutations and resulted in a T–K conversion at codon 3, a G–S at codon 49, an S–N at condon 154 and a T–M at codon 252, respectively. The deletion 187Del24 was identified as a deletion of the second octapeptide repeat (R2) at nucleotides 187–210 (Fig. 1).
Genotype, allele and haplotype frequencies of the PRNP
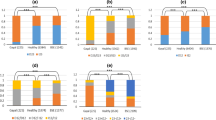
The genotype and gene frequencies of 11 polymorphic sites are summarized in Fig. 2. All sites, except G461A, were consistent with the Hardy–Weinberg equilibrium. Haplotypes covering the 11 loci were identified (Fig. 3). The majority (71.2%, n = 89) of the gayal had the I/I genotype (6:6) in the PRNP N-terminal octapeptide repeat region, 26.4% (n = 33) carried the I/D genotype (6:5) and 2.4% (n = 3) of the animals expressed the D/D genotype (5:5).
Gene homozygosity (Ho), gene heterozygosity (He), effective allele numbers (Ne) and polymorphism information content (PIC) of all loci are shown in Figs. 4 and 5. According to the classification of PIC (low polymorphism for a PIC value <0.25, median polymorphism for a PIC value >0.25 and <0.5, and high polymorphism for a PIC value >0.5) [29], six of eleven loci from the PRNP had a median polymorphic level.
Sequence characteristics of the PRNR and proteins
Compared with bovine PRNP (GenBank accession number AJ298878), the entire coding region of the PRNP from gayal was obtained, which had a 795 bp or 771 bp ORF encoding a protein of 264 or 256 amino acid residues with a predicted molecular mass of 28614 Da (pI 9.40) or 27837 Da (pI 9.40).
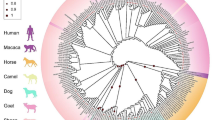
The nucleotide sequence alignments of 20 mammalian PRNP analyses are shown in Fig. 6. The PRNP sequence for gayal shared high identity with the prion protein gene sequences of Zebu (AY720681, 99.4%) and European cattle (EF139165, 99.2%), gaur (AY720697, 99.4%), bison (AY769958, 99.2%), Banteng cattle (AY720693, 98.9%), yak (AY367635, 99.4%), buffalo (AY768533, 98.6%), cape buffalo (AY720686, 97.6%), red deer (AY748455, 97.1%), sika deer (EF057409, 97.4%), fallow deer (EF165089, 97.0%), sheep (DQ149388, 97.9%), goats (EF140716, 97.7%) and cats (AF003087, 97.4%). There also was 80–90% homology with the PRNP sequences for mink (S46825), dogs (EF139170), arctic foxes (EU365392), humans (AF076976) and mice (M18070). Moreover, amino acid alignments for the 20 species revealed higher homology than the nucleotide level (data not shown).
Nucleotide sequences alignments of 20 mammalian prion protein genes. 1. Gayal (Bos frontalis, HQ262495), 2. Zebu cattle (Bos indicus, AY720681), 3. European cattle (Bos taurus, EF139165), 4. Gaur (Bos gaurus, AY720697%), 5. Bison (Bison bison, AY769958), 6. Banteng cattle (Bos jsvanicus, AY720693), 7. Yak (Bos grunniens, AY367635), 8. Buffalo (Bubalus bubalus, AY768533), 9. Cape buffalo (Syncerus caffer, AY720686), 10. Red deer (Cervus elaphus, AY748455), 11. Sika deer (Cervus nippon, EF057409), 12. Fallow deer (Dama dama, EF165089), 13. Sheep (Ovis aries, DQ149388), 14. Goat (Capra hircus, EF140716), 15. Mink (Mustelan vison, S46825), 16. Cat (Felis catus, AF003087), 17. Dog (Canis familiaris, EF139170), 18. Arctic fox (Vulpes lagopus, EU365392), 19. Human (Homo sapiens, AF076976), 20. Mouse (Mus musculus, M18070)
Based on bioinformatic analyses, the signal peptide (SP) was identified from amino acids 1–24 at the N-terminus. An octapeptide repeat region was identified between amino acids 54–103 comprising two nonapeptides (PQ/HGGGGWGQ) surrounding a tandem repeat of three or four copies of an octapeptide (PHGGGWGQ). Three α-helices corresponded to residues 157–163, 184–204, and 211–237, respectively. Two β-sheets were located at residues 136–139 and 178–181. Two N-linked glycosylation sites were located at amino acids 192–194 and 208–210 which represented NIT and NFT, respectively. The putative topology of the gayal PRNP harbored one hydrophobic domain from amino acids 122–145 near the C-terminus. A glycosylphosphatidylinositol (GPI) anchor site between amino acids 241–264 was found. Two cysteine residues (amino acids 190 and 225) involved in the formation of a disulfide bridge were also found (Fig. 7).
The phylogenetic tree analyses revealed that the gayal PRNP had a very close genetic relationship with the PRNP gene of Zebu cattle (Fig. 8).
Discussion
In the present study, the gayal ORF of PRNP was isolated for the first time. Novel polymorphism analyses expanded the collection of known PRNP from animals within the Bos genus. Collectively, 10 SNPs including 6 silent and 4 mis-sense mutations were identified. Comparing with known SNPs of PRNP from European cattle, yak and swamp buffalo (Bubalus bubalis) in China [17, 19, 30, 31], as well as European and Zebu cattle and their hybrids, Banteng cattle (Bos javanicus), American bison, American buffalo (Syncerus caffer nanus), forest buffalo (Syncerus caffer nanus), lowland anoa (Bubalus depressicornis), Asian water buffalo (Bubalus bubalis), nilgai (Boselaphus tragocamelus) in countries other than China [16, 18, 21, 32, 33], three novel SNPs in the coding region including C60T (D20), G145A (G49S) and C756G (T252M), which have not been reported previously in bovid species, were identified.
Eight classical type structural characters, consisting of a SP, a tandem repeat, three α-helices, two β-sheets, a hydrophobic region, two glycosylation sites, a GPI anchor site and two cysteine residues possibly involved in the formation of a disulfide bridge, which were reported as quite conserved elements in mammals [34], were also identified in the present study. However, two non-silent mutations (one T3K and another I252M) were identified in the SP and GPI anchor regions, respectively (Fig. 7).
The SP encodes the determinants for topogenesis and plays a role in targeting proteins to the endoplasmic reticulum [35]. Increased generation of CtmPRNP, which is a topologic form of PRNP formed at the endoplasmic reticulum, is related to the development of transmissible spongiform encephalopathies (TSE) in humans and transgenic mice [36, 37]. Furthermore, the SP was divided into three significant fragments including the n-region (amino acids 1–9), h-region (amino acids 10–16) and c-region (amino acids 17–24). The n-region is a polar and charged domain. Mutation of the n-region often results in the introduction of charged residues and alternations in the topogenesis of PRNP and the introduction of basic residues that decrease the level of CtmPRNP [35].
In the current study, the K3T mutation (reside 3, comprising the positively charged and basic amino acid lysine) within the n-region of SP was converted to the uncharged and aromatic amino acid threonine. Therefore, more attention should be paid to the topogenesis of PRNPs and their expression level in future research. It is noteworthy that this mutation has been reported in the native Chinese southern yellow cattle (B. taurus) [19] but not in the native Chinese northern yellow cattle [17, 30, 31]. In the present report, we also identified this site in gayal from the south west of China. This observation is consistent with a previous report that gayal may have derived genetic material from Chinese southern yellow cattle [24].
The second non-silent mutation G49S was located within the N-terminal region and included amino acids 1–91, comprising the signal peptide (amino acids 1–24) and the tandem repeat region (amino acids 54–103). These play an important role in TSE pathogenesis through possible combination of the N-terminal region with copper ions [38]. Within this area, one linkage is between the octarepeat and the structural domain at His107 and His122 and another is between W60 and G93 in bovine PRNP [39]. However, G49S occurs outside of these regions. Therefore, this mutation G49S may not be related to susceptibility to BSE. Significantly, this site has been identified only in gayal and never in other bovid animals [17, 19–21].
The third non-silent mutation S154 N (analogous to mouse residue 142) is located at the tail of first α-helix [40]. This site was retrieved previously in swamp buffalo and also European and Zebu cattle and their hybrid offspring [17, 19–21]. In transgenic mice carrying chimeric murine–ovine PRNPc, mushp-PRNPcN142S is established after converting asparagine to serine at residue 142. The change leads into a large reduction in Me7 prion-induced conversion, but not the BSE-induced conversion [41]. Serine and asparagine are located on an uncharged polar side chain and serine is considerably smaller than asparagine. Moreover, serine lacks the terminal carboxyamide group which in asparagine enables the formation of hydrogen bonds and improves the intrinsic stability of the molecule [19, 41]. It is possible that this mutation in gayal could lead to the synthesis of a highly stable of prion protein, unlike mushp-PRNPcN142S of transgenic mice, in which a negative result was obtained in a cell-free conversion experiment.
The fourth non-silent mutation, T252M, occurred in the GPI anchor site region which enables the protein prion coded for to associate with sphingolipid- and cholesterol-rich membranes [42] leading to binding and replication [43]. This association is critical for the conformational conversion of normal PRNPc to pathogenic PRNPSc [44, 45]. Only one SNP within the GPI anchor site has been reported in 751 (G or A, V251M) in forest buffalo [21]. In the present study, SNP C756G was predicted to alter the amino acid at 252 from isoleucine to methionine which had not been reported previously. These two amino acids are non-polar but, the functional significance of a T252M variant is unknown.
Turning to the octapeptide repeat, in previous studies it has been found that most healthy cattle from modern breeds have similar distributions of the PRNP octarepeat 6:6 and 6:5 genotypes. This has been found in studies from Japan [46, 47], Korea [48–51], Vietnam, Laos, Myanmar, Mongolia and Bangladesh [52, 53], China [19], USA [54], Croatia [55], Czechoslovakia [56] and Poland [57]. In four comparative studies from Germany [58], Japan [59], Czechoslovakia [26] and Scotland [60] no differences between the frequencies of these PRNP genotypes in healthy and BSE-affected cattle were found. The number of octarepeats in cattle has been found to vary from 4 to 7 [61]. An increased number of octapeptide repetitions in bovine PRNP may be associated with increased host susceptibility to BSE agents [62, 63]. The observed allele frequencies (6, 5 and 4) of octapeptide repeat units were 0.77, 0.14 and 0.09 from 11 gayal samples which were collected in Myanmar where is close to our sampling site near Dulong Town, Gongshan County, Yunnan Province, China [52]. In the current study, based on 125 tested samples, it was found that gayal has a small number of octapeptide repeats, lacking the four, seven or larger number of repeats reported by others for cattle [61]. Furthermore, the genotype and gene frequencies of the PRNP do not differ from those reported above. Therefore, the tandem repeat with R2 del polymorphism in gayal appears not to affect resistance or susceptibility of gayal to BSE.
Recently, a novel E211K mutation (analogous to human reside E200K) was reported to have a relationship with a case of H-type BSE [64–66]. However, this SNP has never been found in gayal or in so-called cattle (including Chinese yellow cattle), swamp buffalo and yak [17, 19, 30, 31].
In conclusion, the present genetic analyses extend knowledge of the DNA polymorphism of the PRNP in gayal. It appears that there is scope for exploring the relationship between the diversity of the coding region for PRNP and resistance to BSE. Further studies should be focused on the Indel polymorphisms in the promoter and intron of this gene.
References
Prusiner SB (1996) Molecular biology and pathogenesis of prion diseases. Trends Biochem Sci 21:482–487
Prusiner SB (1998) Prions. Proc Natl Acad Sci USA 95:13363–13383
Prusiner SB, Scott MR, De Armond SJ, Cohen FE (1998) Prion protein biology. Cell 93:337–348
Aguzzi A (2006) Prion diseases of humans and farm animals: epidemiology, genetics, and pathogenesis. J Neurochem 97:1726–1739
Wang F, Wang X, Yuan CG, Ma J (2010) Generating a prion with bacterially expressed recombinant prion protein. Science 327:1132–1135
Vaccari G, Panagiotidis CH, Acin C, Peletto S, Barillet F, Acutis P, Bossers A, Langeveld J, van Keulen L, Sklaviadis T, Badiola JJ, Andreéoletti O, Groschup MH, Agrimi U, Foster J, Goldmann W (2009) State-of-the-art review of goat TSE in the European Union, with special emphasis on PRNP genetics and epidemiology. Vet Res 40:48
Mondal M, Dhali A, Rajkhowa C, Prakash BK (2004) Secretion patterns of growth hormone in growing captive mithuns (Bos frontalis). Zool Sci 21:1125–1129
Rajkhowa S, Sarma DK, Rajkhowa C (2006) Seroprevalence of toxoplasma gondii antibodies in captive mithuns (Bos frontalis) from India. Vet Parasitol 135:369–374
Chi J, Fu B, Nie W, Wang J, Graphodatsky AS, Yang F (2005) New insights into the karyotypic relationships of Chinese muntjac (Muntiacus reevesi), forest musk deer (Moschus berezovskii) and gayal (Bos frontalis). Cytogenet Genome Res 108:310–316
Mao HM, Deng WD, Wen JK (2005) The biology characteristics of gayal (Bos frontalis) and potential exploitation and utilization. J Yunnan Agric Univ 20:258–261 (In Chinese with English abstract available)
Bhambhani R, Kuspira J (1969) The somatic karyotypes of American bison and domestic cattle. Can J Genet Cytol 11:243–249
Gallagher DS, Womack JE (1992) Chromosome conservation in the Bovidae. J Hered 83:287–298
Zhao KD, Ou CH, Huang YL, He TB (2003) Rare animal germplasm resources in Yunnan Province: Present situation and countermea sures of preservation and research on Dulong cattle (Bos frontalis). J Yellow Cattle Sci 29:71–74 (In Chinese with English abstract available)
Xi DM, Wanapat M, Deng WD, He TB, Yang ZF, Mao HM (2007) Comparison of gayal (Bos frontalis) and Yunnan Yellow Cattle (Bos taurus): in vitro dry matter digestibility and gas production for a range of forages. Asian-Aust J Anim Sci 20:1208–1214
Mohan M, Chandan R, Prakash BS (2007) Development and validation of a highly sensitive economic enzymeimmunoassay for prolactin determination in blood plasma of mithun (Bos frontalis) and its application during milk let down and cyclicity. Anim Reprod Sci 99:182–195
Heaton MP, Leymaster KA, Freking BA, Hawk DA, Smith TP, Keele JW, Snelling WM, Fox JM, Chitko-McKown CG, Laegreid WW (2003) Prion gene sequence variation within diverse groups of U.S. sheep, beef cattle, and deer. Mamm Genome 14:765–777
Zhang L, Li N, Fan B, Fang M, Xu W (2004) PRNP polymorphisms in Chinese ovine, caprine and bovine breeds. Anim Genet 35:457–461
Kues WA, Ollhoff RD, Carnwath JW, de Souza FP, Madeira HMF, Niemann H (2006) High incidence of single nucleotide polymorphisms in the prion protein gene of native Brazilian Caracu cattle. J Anim Breed Genet 123:326–330
Zhao H, Wang XY, Zou W, Zhang YP (2010) Prion protein gene (PRNP) polymorphisms in native Chinese cattle. Genome 53:138–145
Brunelle BW, Greenlee JJ, Seabury CM, Brown CE II, Nicholson EM (2008) Frequencies of polymorphisms associated with BSE resistance differ significantly between Bos taurus, Bos indicus, and composite cattle. BMC Vet Res 4:36
Seabury CM, Honeycutt RL, Rooney AP, Halbert ND, Derr JN (2004) Prion protein gene (PRNP) variants and evidence for strong purifying selection in functionally important regions of bovine exon 3. Proc Natl Acad Sci USA 101:15142–15147
Peletto S, Perucchini M, Acín C, Dalgleish MP, Reid HW, Rasero R, Sacchi P, Stewart P, Caramelli M, Ferroglio E, Bozzetta E, Meloni D, Orusa R, Robetto S, Gennero S, Goldmann W, Acutis PL (2009) Genetic variability of the prion protein gene (PRNP) in wild ruminants from Italy and Scotland. J Vet Sci 10:115–120
Wang Y, Qin Z, Bao Y, Qiao J, Yang L, Zhao D (2009) Sequence analysis of the prion protein gene in Mongolian gazelles (Procapra gutturosa). Virus Genes 39:273–278
Gou X, Wang YQ, Yang SL, Deng WD, Mao HM (2010) Genetic diversity and origin of gayal and cattle in Yunnan revealed by mtDNA D-loop and SRY gene sequence variation. J Anim Breed Genet 127:154–160
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, New York
Hresko S, Mojzis M, Tkacikova L (2009) Prion protein gene polymorphism in healthy and BSE-affected Slovak cattle. J Appl Genet 50:371–374
Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleaes. Proc Natl Acad Sci USA 76:5269–5273
Zhao M, Chen H, Wang X, Yu H, Wang M, Wang J, Lan XY, Zhang CF, Zhang LZ, Guo YK, Zhang B, Hu SR (2009) aPCR-SSCP and DNA sequencing detecting two silent SNPs at KAP8.1 gene in the Cashmere goat. Mol Biol Rep 36:1387–1391
Wu R, Xie QG, Liu XT, Chen HT (2004) Cloning and sequence analysis of the Bovine prion protein (bPrPc) gene. Acta Vet Zootech Sin 35:318–332 (In Chinese with English abstract available)
Qin LH, Zhao YM, Bao YH, Bai WL, Chong J, Zhang GL, Zhang JB, Zhao ZH (2010) Polymorphism of the prion protein gene (PRNP) in two Chinese indigenous cattle breeds. Mol Biol Rep. doi:10.1007/s11033-010-0541-0
Takasuga A, Abe T, Ito T, Watanabe T, Kamatani N, Sugimoto Y (2003) Novel prion protein polymorphisms in cattle. Anim Genet 34:396–397
Sakudo A, Xue G, Kawashita N, Ano Y, Takagi T, Shintani H, Tanaka Y, Onodera T, Ikuta K (2010) Structure of the prion protein and its gene: an analysis using bioinformatics and computer simulation. Curr Protein Pept Sci 11:166–179
Wopfner F, Weidenhofer G, Schneider R, von Brunn A, Gilch S, Schwarz TF (1999) Analysis of 27 mammalian and 9 avian PrPs reveals high conservation of flexible regions of the prion protein. J Mol Biol 289:1163–1178
Kim SJ, Rahbar R, Hegde RS (2001) Combinatorial control of prion protein biogenesis by the signal sequence and transmembrane domain. J Biol Chem 276:26132–26140
Hegde RS, Mastrianni JA, Scott MR, DeFea KA, Tremblay P, Torchia M, DeArmond SJ, Prusiner SB, Lingappa VR (1998) A transmembrane form of the prion protein in neurodegenerative disease. Science 279:827–834
Hegde RS, Tremblay P, Groth D, DeArmond SJ, Prusiner SB, Lingappa VR (1999) Transmissible and genetic prion diseases share a common pathway of neurodegeneration. Nature 402:822–826
Lawson VA, Priola SA, Meade-White K, Lawson M, Chesebro B (2004) Flexible N-terminal region of prion protein influences conformation of protease-resistant prion protein isoforms associated with cross-species scrapie infection in vivo and in vitro. J Biol Chem 279:13689–13695
Viles JH, Klewpatinond M, Nadal RC (2008) Copper and the structural biology of the prion protein. Biochem Soc Trans 36:1288–1292
López García F, Zahn R, Riek R, Wüthrich K (2000) NMR structure of the bovine prion protein. Proc Natl Acad Sci USA 97:8334–8339
Kupfer L, Eiden M, Buschmann A, Groschup MH (2007) Amino acid sequence and prion strain specific effects on the in vitro and in vivo convertibility of ovine/murine and bovine/murine prion protein chimeras. Biochim Biophys Acta 1772:704–713
Baron BS, Caughey B (2003) Effect of glycosylphosphatidylinositol anchor-dependent and -independent prion protein association with model raft membranes on conversion to the protease-resistant isoform. J Biol Chem 278:14883–14892
Bate C, Tayebi M, Williams A (2010) The glycosylphosphatidylinositol anchor is a major determinant of prion binding and replication. Biochem J 428:95–101
Taraboulos A, Scott M, Semenov A, Avrahami D, Laszlo L, Prusiner SB (1995) Cholesterol depletion and modification of COOH-terminal targeting sequence of the prion protein inhibit formation of the scrapie isoform. J Cell Biol 129:121–132
Baron T (2002) Mouse models of prion disease transmission. Trends Mol Med 8:495–500
Abe T, Hasebe H, Kobayashi E (2006) Frequencies of bovine prp gene polymorphisms in Holstein and Japanese Holstein and Japanese black bulls in Japan. Anim Sci J 77:395–398
Msalya G, Shimogiri T, Nishitani K, Okamoto S, Kawabe K, Minesawa M, Maeda Y (2010) Indels within promoter and intron 1 of bovine prion protein gene modulate the gene expression levels in the medulla oblongata of two Japanese cattle breeds. Anim Genet 41:218–221
Jeong BH, Lee YJ, Kim NH, Carp RI, Kim YS (2006) Genotype distribution of the prion protein gene (PRNP) promoter polymorphisms in Korean cattle. Genome 49:1539–1544
Jeong BH, Sohn HJ, Lee JO, Kim NH, Kim JI, Lee SY, Cho IS, Joo YS, Carp RI, Kim YS (2005) Polymorphisms of the prion protein gene (PRNP) in Hanwoo (Bos taurus coreanae) and Holstein cattle. Genes Genet Syst 80:303–308
Choi SH, Chae SH, Choi HH, Kim JS, Kang BR, Yeo JS, Choi I, Lee YS, Choy YH, Park HS (2007) Genomic sequence variability of the prion gene (PRNP) in Korean cattle. Asian-Aust J Anim Sci 20:653–660
Kim Y, Kim JB, Sohn H, Lee C (2009) A national survey on the allelic, genotypic, and haplotypic distribution of PRNP insertion and deletion polymorphisms in Korean cattle. J Genet 88:99–103
Shimogiri T, Msalya G, Myint SL, Okamoto S, Kawabe K, Tanaka K, Mannen H, Minezawa M, Namikawa T, Amano T, Yamamoto Y, Maeda Y (2010) Allele distributions and frequencies of the six prion protein gene (PRNP) polymorphisms in Asian native cattle, Japanese breeds, and mythun (Bos frontalis). Biochem Genet 48:829–839
Muramatsu Y, Sakemi Y, Horiuchi M, Ogawa T, Suzuki K, Kanameda M, Hanh TT, Tamura Y (2008) Frequencies of PRNP gene polymorphisms in Vietnamese dairy cattle for potential association with BSE. Zoonoses Public Health 55:267–273
Clawson ML, Heaton MP, Keele JW, Smith TP, Harhay GP, Laegreid WW (2006) Prion gene haplotypes of U.S. cattle. BMC Genet 7:51
Premzl M, Bozic P, Gamulin V (2000) PRNP octarepeat allele genotype frequencies among the modern and rare cattle breeds in Croatia. Anim Genet 31:408–409
Tkacikova A, Voralek R, Filipaik P, Mikula I Sr (2007) Octapeptide polymorphism analysis of Slovak autochthonous cattle breeds. Acta Vet Brno 76:47–50
Walawski K, Czarnik U (2003) Prion octapeptide-repeat polymorphism in Polish Black-and-White cattle. J Appl Genet 44:191–195
Sander P, Hamann H, Pfeiffer I, Wemheuer W, Brenig B, Groschup MH, Ziegler U, Distl O, Leeb T (2004) Analysis of sequence variability of the bovine prion protein gene (PRNP) in German cattle breeds. Neurogenetics 5:19–25
Nakamitsu S, Miyazava T, Horuichi M, Onoe S, Ohoba Y, Kitagawa H, Ishiguro N (2006) Sequence variation of bovine prion protein gene in Japanese cattle (Holstein and Japanese Black). J Vet Med Sci 68:27–33
Hunter N, Goldmann W, Smith G, Hope J (1994) Frequencies of PrP gene variants in healthy cattle and cattle with BSE in Scotland. Vet Rec 135:400–403
Goldmann W (2008) PrP genetics in ruminant transmissible spongiform encephalopathies. Vet Res 39:30
Castilla J, Gutiérrez-Adán A, Brun A, Pintado B, Parra B, Ramírez MA, Salguero FJ, Díaz San Segundo F, Rábano A, Cano MJ, Torres JM (2004) Different behavior toward bovine spongiform encephalopathy infection of bovine prion protein transgenic mice with one extra repeat octapeptide insert mutation. J Neurosci 24:2156–2164
Castilla J, Gutiérrez-Adán A, Brun A, Pintado B, Salguero FJ, Parra B, Segundo FD, Ramírez MA, Rábano A, Cano MJ, Torres JM (2005) Transgenic mice expressing bovine PrP with a four extra repeat octapeptide insert mutation show a spontaneous, non-transmissible, neurodegenerative disease and an expedited course of BSE infection. FEBS Lett 579:6237–6246
Heaton MP, Keele JW, Harhay GP, Richt JA, Koohmaraie M, Wheeler TL, Shackelford SD, Casas E, King DA, Sonstegard TS, Van Tassell CP, Neibergs HL Jr, Chase CC, Kalbfleisch TS, Smith TP, Clawson ML, Laegreid WW (2008) Prevalence of the prion protein gene E211K variant in U.S. cattle. BMC Vet Res 4:25
Nicholson EM, Brunelle BW, Richt JA, Kehrli ME Jr, Greenlee JJ (2008) Identification of a heritable polymorphism in bovine PRNP associated with genetic transmissible spongiform encephalopathy: evidence of heritable BSE. PLoS ONE 3:e2912
Richt JA, Hall SM (2008) BSE case associated with prion protein gene mutation. PLoS Pathog 4:e1000156
Acknowledgments
Major funding was provided from the State Key Laboratory of Veterinary Etiological Biology, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences (SKLVEB2009KFKT010) and in part funding was also provided by the Open-funded Project (to DMX) of the Faculty of Animal Science and Technology, Yunnan Agricultural University. We thank to our team members for sharing and using samples from gayal which were collected between 2005 and 2008 with financial support from the Natural Science Foundation of Yunnan Province (2005C0038M), the National “863” Program (2008AA101001) and the National Technology Support Project (2008BADB2B01-14). We acknowledge with gratitude the financial support provided. The authors thank Professor G. H. McDowell for invaluable comments on the manuscript. The authors also appreciated the help of staff from the Gongshan County Animal Science and Veterinary Bureau during samples collection.
Author information
Authors and Affiliations
Corresponding author
Additional information
D. Xi and Q. Liu contributed equally to this work and should be considered co-first authors.
Rights and permissions
About this article
Cite this article
Xi, D., Liu, Q., Guo, J. et al. Genetic variability of the coding region for the prion protein gene (PRNP) in gayal (Bos frontalis). Mol Biol Rep 39, 2011–2020 (2012). https://doi.org/10.1007/s11033-011-0948-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-0948-2