Abstract
Killer cell immunoglobulin-like receptors (KIRs) are a family of inhibitory and activating receptors expressed by natural killer (NK) cells and regulate NK cells’ activity. KIR genes are highly polymorphic markers, characterized by a wide diversity, and can therefore be considered as good population genetic markers. The aim of this study was to determine KIR gene frequencies, ratios of haplotypes and genotypes in Southern Turkey and also to compare the data with other worldwide populations studied previously. The study group consisted of 200 non-related individuals from Southern Turkey. The percentage of each KIR gene in the population group was determined by direct counting. Differences between populations in the distribution of each KIR gene and genotype profile were estimated by two-tailed Fisher Exact test. The most frequent non-framework KIR genes detected in Southern Turkey population were: KIR 2DL1 (97%), KIR 3DL1 (91%), KIR 2DS4 (92%) and the pseudogene 2DP1 (96%). Fourty different genotypes were found in 200 subjects and AA1 genotype was the most frequent (27%). Among 40 different genotypes, ten of these were described for the first time in this study and were added to the database (http://www.allelefrequencies.net) numerized as genotype ID from 400 to 409. Gene frequencies and found genotypes demonstrated similarity of Southern Turkey’s KIR repertoire with the KIR repertoires of Middle East and European population. High variability seen in KIR genome in this region is thought to be formed as a result of migration and settlement of different civilizations in this region and heterogenity formed in time.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Natural killer (NK) cells have important immunoregulatory functions in the innate immune response such as control of tumour cells, virally infected cells and rejection of allogeneic bone marrow transplants [1–4]. Effector functions of NK cells are regulated by a subtle balance between the signals transduced from activating and inhibitory cell-surface receptors [5, 6]. Killer cell immunoglobulin-like receptors (KIRs) are members of the group of cell-surface molecules that activate or inhibit NK cells’ interaction with human leukocyte antigen (HLA) class I molecules on the surface of target cells [3, 5, 7].
KIRs are glycoproteins classified according to their structure and function. Each KIR molecule consists of two or three extracellular immunoglobulin domains (2D and 3D molecules, respectively), a transmembrane part and a short (S) or a long (L) intracellular tail [8–11]. Up to date, Sixteen KIR genes have been characterized [7]. Fourteen of them encode receptors triggering either inhibition (3DL1–3, 2DL1–3, 2DL5) or activation (3DS1, 2DS1–5) or both (2DL4), and two pseudogenes (2DP1 and 3DP1) do not encode cell-surface receptors. Inhibitory KIRs (iKIRs) recognize distinct motifs of the polymorphic HLA class I molecules and trigger signals that stop effector function of NK cells [7]. The healthy cells are protected from NK cell surveillance by expressing HLA class I molecules. If the cells lose their HLA class I expression after viral infections or tumor transformation, they become potential targets for NK cell response. On the other hand an intriguing model proposed from genetic association studies is that the signals transduced by activating KIRs (aKIRs) upon their interaction with putative ligands expressed on the surface of stressed cell targets (i.e., virally infected or tumor transformed cells) overcame iKIR+HLA-mediated inhibition and trigger NK cell response against the nonself (leads to control of infection) or self (leads to autoimmunity) [12, 13].
KIR gene family is located within the leucocyte receptor cluster on chromosome 19q13.4. [3, 7, 14]. Three genes; 2DL4, 3DL2, 3DL3 and one pseudogene 3DP1 are present in virtually all KIR haplotypes and are termed framework loci [3, 10]. According to the order of KIR genes and gene content, haplotypes are divided into A and B. The A haplotype contains six loci coding inhibitory receptors 2DL1, 2DL3, 2DL4, 3DL1, 3DL2, 3DL3, and only one activating KIR 2DS4. The B haplotype is presented by a great variety of subtypes that differ from each other mostly by combinations of stimulatory receptors [10]. As the number of KIR genes in different haplotypes varies, a wide range of genotypes in different ethnic populations may be observed [13, 15, 16]. Sixteen distinct KIR gene loci which vary with respect to their presence or absence on different KIR haplotypes, creating considerable diversity in the number or KIR genotypes were observed in the population. Allelic polymorphism at individual KIR genes also exists, further contributing to the level of KIR diversity observed [3].
From a functional point of view, KIR gene diversity could explain the differential impact of KIR markers in many clinical fields. Thus, KIR diversity do not only explain susceptibility to viral infections and to tumour development but could also explain situations in haematopoietic stem cell transplantation where particular KIR and HLA ligand mismatches lead to graft versus leukaemia and/or acute graft versus host disease [17]. An extensive study of KIR diversity could help us to better understand mechanisms of NK lysis related to this diversity.
Based on the KIR genome diversity, the world populations can be divided into six broad groups, roughly corresponding to continents: Africans, Northeast Asians, Mexicans, American Natives, Asian Indians, and Europeans. These populations emerged from at least three major waves of prehistoric human migrations from Africa [13, 18]. There has been an explosion in population studies determining the frequency of KIR genes. However, there is still limited knowledge of allele and haplotype frequencies in different populations. Therefore, this study was planned as a population genetic study due to the lack of data on KIR gene frequencies, ratios of haplotypes and genotypes in Southern Turkey. To our knowledge, no analogous population genetic study has been performed in Turkey so far.
Materials and methods
Study subjects and samples
The study group consisted of 200 individuals (111 female, 89 male) convenient for a population study from Southern Turkey that locates at geographical coordinates 36°47′51′′ North and 34°37′48′′ East. This region known as Anatolia has been inhabited since the ninth millennium BC. A fortification was put up around 4500 BC, but the site appears to have been abandoned between 3200 and 1200 BC. In the following centuries this region became a part of many states and civilizations including the Hittites, Assyrians, Persians, Greeks, the Macedonians of Alexander the Great, Seleucids, Lagids, Romans, Arabs, Egyptian Tulunids, Seljuk Turks, Mongols, Crusaders, Armenians, Mamluks, Anatolian beyliks and Ottomans [20]. It was liberated by the Turkish Republic in 1920. The official language of this area is Turkish.
All individuals included to study were healthy, unrelated and randomly selected. All of these individuals’ three generation ancestors were born and lived in Southern Turkey. Peripheral blood samples were collected after informed consent and according to the permit given by the Ethical Review Board of the Medical Faculty of Mersin University. Genomic DNA was extracted from whole blood samples using DNA isolation kit (Qiagen EZ1, cat no: 951034, QIAGEN Vertriebs GmbH, Vienna, Austria) using Genovision Geno M-6 Biorobot technology (GenoVision Inc. Philadelphia, USA).
KIR genotyping
Genotyping of the 16 KIR genes was performed using the multiplex KIR-SSO typing kit from Tepnel Lifecodes Corporation (Ref: 545110, Connecticut, USA). This product consists of a mixture of locus-specific oligonucleotide probes coupled to color-coded microspheres (Luminex Corp) and two PCR reactions for the amplification of KIR-exons 4, 5, 7, 8, and 9. To type each sample, the PCR was performed and the product was hybridized with the SSO-probe mixture using the manufacturer’s protocol. After hybridization, the sample plate was placed in a Luminex instrument for analysis.
Prediction of group-A and -B KIR haplotypes
Frequencies of group-A and -B KIR haplotypes were deduced from the genotype data [21]. Individuals carrying only KIR3DL3, 2DL3, 2DL1, 2DP1, 3DP1, 2DL4, 3DL1, 2DS4, 3DL2, a fixed gene content characteristic of group-A haplotypes, were considered carrying two copies of group-A KIR haplotypes (AA genotypes) (Middleton et al. 2003) [21]. If any of genes KIR2DL2, 2DL5, 3DS1, 2DS1, 2DS2, 2DS3, 2DS5 were present genotype is taken as having B (Bx).
Data analysis and statistical methods
The percentage of each KIR gene in the population group was determined by direct counting (individuals positive for the gene/individuals tested per population × 100). The frequency data of KIR genes and genotypes in 13 other populations to be compared to Southern Turkey population was extracted from the selected publications and from the ‘http://www.allelefrequencies.net’ database [15, 16, 21–30]. Differences between populations in the distribution of each KIR gene and genotype profile were estimated by two-tailed Fisher Exact test and P < 0.05 was considered to be statistically significant [15].
Results
Our results showed that KIR gene frequencies of the population who lives in Southern Turkey are highly variable (Fig. 1). KIR genes of Southern Turkey population shows a higher frequency of inhibitory receptors than activating ones. The ratios of the framework genes were found 99.5% for KIR3DL3, 99% for KIR2DL4, 99.5% for KIR3DL2 and 99% for pseudogene KIR3DP1 (Fig. 1). The most frequent non-framework KIR genes detected in Southern Turkey population were: KIR2DL1 (97%), KIR3DL1 (91%), KIR2DS4 (92%) and the pseudogene 2DP1 (96%) (Fig. 1). The rarest KIR gene in the Southern Turkey population was 2DS3 which was found in 33% of genotypes. Since the methods used for KIR genotyping permitted to distinguish groups of alleles of KIR 2DS4, we found that 2DS4*001–002 were present in 31.4% of individuals, while alleles 2DS4*003–006 were found in 68.6%. The frequency of A haplotypes (50.5%) were equally distributed as compared to the group-B haplotypes (49.5%) with an A:B ratio of 1.02:1.
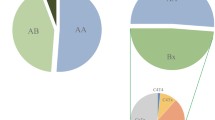
Fourty different genotypes were found in 200 unrelated Turkish individuals. Among 40 different genotypes, ten of these do not exist in the KIR genotype database [21] and were found for the first time in this study (Fig. 2). These genotypes were added to database numerized as genotype ID from 400 to 409 [21] (Table 1). The most frequent genotype found in 27% of individuals was 2DL1, 2DL3, 2DL4, 2DS4, 3DL1, 3DL2, 3DL3, 2DP1, 3DP1 which corresponds to A haplotype homozygosity (AA1) (Fig. 2). Also, three more rare AA genotypes were found (two of them were firstly found in this study): in two individuals KIR 2DL3 was absent in addition to the described set of genes earlier (AA180), in one individual KIR 2DP1 and 3DP1 were absent (new genotype, AA409) and in one individual 3DL3 was absent (new genotype, AA407). Other analysed individuals demonstrated the presence of more than one activating gene and thus, may be considered as Bx. Six individuals from the analysed cohort possessed all 16 KIR genes known today. The remaining genotypes contained between 8 and 15 KIR genes. Seventeen genotypes occurred only once (Fig. 2).
KIR genotype profiles of Southern Turkey population. Fourty genotypes that differed from each other by the presence (shaded box) and absence (white box) of 16 KIR genes were observed. The figure consisted the percentage frequency and defined the number of individuals carrying the genotype. Genotype ID refers to genotype classification according to http://www.allelefrequencies.net
In comparison to other populations, it was remarkable that the KIR gene frequencies of Southern Turkey Population were quite similar to Lebanese population that there is no statistically significant difference between all KIR genes (P < 0.05) [27]. The frequencies of 2DL1 and 2DS5 (which is a characteristic gene of B haplotype) genes in Southern Turkey population were higher than that in Palestinian (P < 0.001, P = 0.041) [27] and Greek (P = 0.007, P = 0.002) [16] populations which lies on the same geographic location (Table 1). In addition, The frequencies of only two genes (2DL3 and 2DS5) of our population were higher than French population and there is no statistically difference between other genes [15]. In comparison to German population there were statistically significant differences between 2DL5, 2DS4, 2DS5 genes (P = 0.027, P = 0.041, P = 0.029) [29]. In addition, when we compare our gene frequencies with the gene frequencies of Northern Europian (Finish and Irish) populations the main differences were seen especially in 2DL3 genes (P < 0.001, P = 0.032) [15, 24] (Table 1).
Compared to East Asian populations, the frequencies of 2DL2, 2DL5, 2DS2 and 2DS3 genes in Southern Turkey population were higher than that in Chinese and Japanese populations, while the frequencies of 2DL3 gene were lower [23, 30] (Table 1). In addition, main differences in the distribution of some KIR genes were also observed when our population was compared to Indian, Senegal, Mexican populations and Australian Aborgins [15, 22, 26, 28] (Table 1).
Discussion
Genes coding the main molecules involved in the human immune system—immunoglobulins, HLA molecules and KIRs—exhibit a very high level of polymorphism that reveals remarkable frequency variations in human populations. It is now widely accepted that KIR genes are co-evolving with their HLA ligands [31]. In addition, genotypic KIR variability is also a consequence of allelic and haplotypic diversity and results in the fact that two unrelated individuals with identical KIR genotypes are difficult to find [10]. With these characteristic features, KIR genes can be considered as good population genetic markers [15, 16, 27].
Turkey which lies on a central geographic location between Europe and Asia, and has extended period of human occupation, and a hierarchical social structure, provides unique opportunities for exploring the diversity of the rapidly evolving KIR genes. This research is the first population study focused on characterizing KIR genes in Turkish population. As a result of our study, genes KIR2DL1, 2DL3, 2DL4, 3DL1, 3DL2, 3DL3, and 2DS4 and the pseudogenes KIR3DP1 and 2DP1 were detected in high frequency. Nevertheless, the activating genes KIR2DS1–3, 2DS5, and 3DS1, and the frequency of the inhibitory receptors KIR2DL2 and 2DL5 showed a greater variability. These data are similar to those found among Caucasians [25, 28, 29]. There is also a case–control study performed in Turkish leukaemia patients by Middleton et al. which showed the distrubition of KIR genes and their ligands. They proposed a model in chronic myeloid leukaemia of protection via KIR2DL2 and/or KIR2DS2 with the presence of the ligand HLA-C1 group and susceptibility via HLA-Bw4 homozygosity [32].
Comparing our results with the study of the KIR gene repertoire in different populations, we found some basic genotypic features and very similar gene frequencies with the Lebanese, French, Greek, Phalestinian, German and Finnish populations [15, 16, 25, 27, 29]. However, some major differences were observed in comparison to East Asian [23, 30], South Asian and Australian aboriginal populations [26, 33]. Compared to East Asian populations, the frequencies of 2DL2 and 2DL5 genes in Southern Turkey population were higher than that in Chinese and Japanese populations, while the frequencies of 2DL3 gene were lower [23, 30]. The frequencies of KIR2DS2 and 2DS3 were significantly different from that reported in Australian Aborigines and from what has been described for East Asian populations [23, 28, 34]. In Southern Turkey Population, KIR2DS4 receptor was predominantly represented by the non-expressed allelic form KIR1D comprising KIR2DS4*003/004/006/007 alleles [35]. According to our results, similarities exist in KIR genes between our region, Middle East and European countries which were all affected from the third migration wave by prehistoric human migration and which lies on the same geographic location.
The content of KIR genes was grouped into genotypes and more than 380 genotypes have been identified up to date [21]. We found 40 genotypes in the Southern Turkey population. Thirty of these genotypes were also found in other populations. Among 40 different genotypes that we determined, 10 of these did not exist in the KIR genotype database [21] and were found for the first time in this study. These genotypes were added to the database numerized as genotype ID from 400 to 409. As a result of our study, AA1 genotype was the most frequent (27%) genotype. We also found two new genotypes of AA (AA 407 and AA 409) group. In the Bx group, the most frequent genotypes were Bx2 (10%) and Bx4 (10%) which both existed in 20 of 200 individuals. In addition we found eight new Bx group (Bx400, Bx401, Bx402, Bx403, Bx404, Bx405, Bx406, Bx408) in our population [21].
Previous studies showed that the most frequent genotype was AA1 genotype in all worldwide populations. The percentage of AA1 genotype were 58.7% in Chinese, 56% in Japanese, 15.7% in Southern Asian, 35.3% in African, 1.7% in Australian aborgins and 28.3% in Caucasions [23, 30]. Our percentage of 27% is similar to those found among Caucasians.
Based on the KIR genome diversity, the world populations can be divided into six broad groups, roughly corresponding to continents: Africans, Northeast Asians, Mexicans American Natives, Asian Indians, and Europeans. These populations emerged from at least three major waves of prehistoric human migrations from Africa [13, 18, 19]. Migration-1 was considered to be the first human dispersal out of Africa ~40,000–60,000 years ago, most likely via the tropical coast of Saudi Arabia, India, Southeast Asia, and Australia. Migration-2 occurred via central Asia, China, and, from there, the recent colonization of the American by southward migration across the Behring land bridge ~20,000 years ago. Migration-3 which effected our population occurred in Europe ~40,000 years ago [13]. Groups-A and -B haplotypes are equally distributed in Caucasians suggestive of balancing selection [15, 16, 24, 29, 34]. Conversely, predominance of one haplogroup over the other was observed in some populations. Group-A haplotype was predominant in Northeast Asians (Han Chinese, Korean, and Japanese) which were effected from the earlier stage of migration-2 [23, 34, 36]. The populations emerged from the latter stage of this migration (Native American populations) posses dominant group-B KIR haplotypes [37–39]. In addition group-B haplotype was frequent in North Indians, Pakistani, and Natives of Australia [26, 28, 33]. Overall, the groups-A and -B KIR haplotypes are equally distributed in the populations that emerged from third migration wave which was also thought to effect Turkey population [13]. Our data were consistent with previous studies of these populations which effected from the third migration wave concerning equally distribution of the A haplotype group with the B haplotype group in Southern Turkey population.
As a conclusion, high variability seen in KIR genome in Southern Turkey population is thought to be formed as a result of migration and settlement of different civilizations in this region and heterogenity formed in time. This study may serve as a basis for further analysis of the KIR genes diversity and for the role played by KIR genotypes in transplantation and disease. Recognizing the genetic characteristics of the NK cell KIR receptor can help to understand the biologic function of NK cells at the molecular level. This can allow a better understanding of the special functions and regulatory mechanisms of NK and T cells. Undoubtedly, further functional studies would be required for the precise understanding the complexity of KIRs and the immune mechanisms involving KIRs.
References
French AR, Yokoyama WM (2003) Natural killer cells and viral infections. Curr Opin Immunol 15:45–51
Long EO (2002) Tumour cell recognition by natural killer cells. Semin Cancer Biol 12:57–61
Middleton D, Williams F, Halfpenny IA (2005) KIR genes. Transpl Immunol 14:135–142
Parham P, McQueen KL (2003) Alloreactive killer cells: hindrance and help for haematopoietic transplants. Nat Rev Immunol 3:108–122
Moretta L, Biassoni R, Bottino C, Mingari MC, Moratta A (2000) Human NK-cell receptors. Immunol Today 9:420–422
Lanier LL (2005) NK cell recognition. Annu Rev Immunol 23:225–274
Vilches C, Parham P (2002) KIR: diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol 20:217–251
Lanier LL, Corliss BC, Wu J, Leong C, Phillips JH (1998) Immunoreceptor DAP12 bearing a tyrosine-based activation motif is involved in activating NK cells. Nature 391:703
Olcese L, Cambiaggi A, Semenzato G, Bottino C, Moretta A, Vivier E (1997) Human killer cell activatory receptors for MHC class I molecules are included in a multimeric complex expressed by natural killer cells. J Immunol 158:5083
Pavlova Y, Kolesar L, Striz I, Jabor A, Slavcev A (2008) Distribution of KIR genes in the Czech population. Int J Immunogenet 35:57–61
Moretta L, Moretta A (2004) Killer immunoglobulin-like receptors. Curr Opin Immunol 16:626
Khakoo SI, Carrington M (2006) KIR and disease: a model system or system of models? Immunol Rev 214:186–201
Rajalingam R, Du Z, Meenagh A, Luo L, Kavitha VJ, Pavithra-Arulvani R, Vidhyalakshmi A, Sharma SK, Balazs I, Reed EF, Pitchappan RM, Middleton D (2008) Distinct diversity of KIR genes in three southern Indian populations: comparison with world populations revealed a link between KIR gene content and pre-historic human migrations. Immunogenetics 60:207–217
Wilson MJ, Torkar M, Haude A, Milne S, Jones T, Sheer D, Beck S, Trowsdale J (2000) Plasticity in the organization and sequences of human KIR/ILT gene families. Proc Natl Acad Sci USA 97:4778
Denis L, Sivula J, Gourraud PA, Kerdudou N, Chout R, Ricard C, Moisan JP, Gagne K, Partanen J, Bignon JD (2005) Genetic diversity of KIR natural killer cell markers in populations from France, Guadeloupe, Finland, Senegal and Re′ union. Tissue Antigens 66:267–276
Niokou D, Spyropoulou-Vlachou M, Darlamitsou A, Stavropoulos-Giokas C (2003) Distribution of killer cell immunoglobulin-like receptors in the Greek population. Hum Immunol 64:1167–1176
Ruggeri L, Capanni M, Urbani E et al (2002) Effectiveness of donor natural killer cell alloreactivity in mismatched hematopoietic transplants. Science 295:2097–2100
Goebel T (2007) Anthropology. The missing years for modern humans. Science 315:194–196
Mellars P (2006) Going east: new genetic and archaeological perspectives on the modern human colonization of Eurasia. Science 313:796–800
Mitchell S (1995) Anatolia: land, men, and Gods in Asia minor. Oxford University Press, Oxford, p 41
Middleton D, Menchaca L, Rood H, Komerofsky R (2003) New allele frequency database:http://www.allelefrequencies.net. Tissue Antigens 61:403–407
Gutiérrez-Rodríguez ME, Sandoval-Ramírez L, Díaz-Flores M, Marsh SG, Valladares-Salgado A, Madrigal JA, Mejía-Arangure JM, García CA, Huerta-Zepeda A, Ibarra-Cortés B, Ortega-Camarillo C, Cruz M (2006) KIR gene in ethnic and Mestizo populations from Mexico. Hum Immunol 67:85–93
Jiang K, Zhu FM, Lv QF, Yan LX (2005) Distribution of killer cell immunoglobulin-like receptor genes in the Chinese Han population. Tissue Antigens 65(6):556–563
Middleton D, Meenagh A, Gourraud PA (2007) KIR haplotype content at the allele level in 77 Northern Irish families. Immunogenetics 59(2):145–158
Norman PJ, Stephens HA, Verity DH, Chandanayingyong D, Vaughan RW (2001) Distribution of natural killer cell immunoglobulin-like receptor sequences in three ethnic groups. Immunogenetics 52:195
Rajalingam R, Krausa P, Shilling HG, Stein JB, Balamurugan A, McGinnis MD, Cheng NW, Mehra NK, Parham P (2002) Distinctive KIR and HLA diversity in a panel of north Indian Hindus. Immunogenetics 53(12):1009–1019
Rayes R, Bazarbachi A, Khazen G, Sabbagh A, Zaatari G, Mahfouz R (2007) Natural killer cell immunoglobulin-like receptors (kir) genotypes in two Arab populations: will KIR become a genetic landmark between nations? Immunogenetics 59:145–158
Toneva M, Lepage V, Lafay G, Dulphy N, Busson M, Lester S, Vu-Trien A, Michaylova A, Naumova E, McCluskey J, Charron D (2001) Genomic diversity of natural killer cell receptor genes in three populations. Tissue Antigens 57(4):358–362
Uhrberg M, Parham P, Wernet P (2002) Definition of gene content for nine common group B haplotypes of the Caucasoid population: KIR haplotypes contain between seven and eleven KIR genes. Immunogenetics 54(4):221–229
Yawata M, Yawata N, McQueen KL, Cheng NW, Guethlein LA, Rajalingam R, Shilling HG, Parham P (2002) Predominance of group A KIR haplotypes in Japanese associated with diverse NK cell repertoires of KIR expression. Immunogenetics 54(8):543–550
Sanchez-Mazas A, Fernandez-Viña M, Middleton D, Hollenbach JA, Buhler S, Di D, Rajalingam R, Dugoujon JM, Mack SJ, Thorsby E (2011) Immunogenetics as a tool in anthropological studies. Immunology 133:143–164
Middleton D, Diler AS, Meenagh A, Sleator C, Gourraud PA (2009) Killer immunoglobulin-like receptors (KIR2DL2 and/or KIR2DS2) in presence of their ligand (HLA-C1 group) protect against chronic myeloid leukaemia. Tissue Antigens 73:553–560
Norman PJ, Carrington CV, Byng M, Maxwell LD, Curran MD, Stephens HA, Chandanayingyong D, Verity DH, Hameed K, Ramdath DD, Vaughan RW (2002) Natural killer cell immunoglobulin- like receptor (KIR) locus profiles in African and South Asian populations. Genes Immun 3:86–95
Yawata M, Yawata N, Abi-Rached L, Parham P (2002) Variation within the human killer cell immunoglobulin-like receptor (KIR) gene family. Crit Rev Immunol 22:463–482
Maxwell LD, Wallace A, Middleton D, Curran MD (2002) A common KIR2DS4 deletion variant in the human that predicts a soluble KIR molecule analogous to the KIR1D molecule observed in the rhesus monkey. Tissue Antigens 60:254–258
Whang DH, Park H, Yoon JA, Park MH (2005) Haplotype analysis of killer cell immunoglobulin-like receptor genes in 77 Korean families. Hum Immunol 66:146
Ewerton PD, Leite Mde M, Magalhaes M, Sena L, Melo dos Santos EJ (2007) Amazonian Amerindians exhibit high variability of KIR profiles. Immunogenetics 59:625–630
Flores AC, Marcos CY, Paladino N, Capucchio M, Theiler G, Arruvito L, Pardo R, Habegger A, Williams F, Middleton D, Fainboim L (2007) KIR genes polymorphism in Argentinean Caucasoid and Amerindian populations. Tissue Antigens 69:568–576
Gendzekhadze K, Norman PJ, Abi-Rached L, Layrisse Z, Parham P (2006) High KIR diversity in Amerindians is maintained using few gene-content haplotypes. Immunogenetics 58:474–480
Acknowledgments
This study was approved and funded by BAP of Mersin University, Mersin, Turkey (grant number: BAP-TF-TB(ÖGÖ)2008-3TU).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ozturk, O.G., Polat, G. & Atik, U. Diversity of killer cell immunoglobulin-like receptor genes in Southern Turkey. Mol Biol Rep 39, 1989–1995 (2012). https://doi.org/10.1007/s11033-011-0945-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-0945-5