Abstract
Recently, a C>T polymorphism (rs1434536) in a miR-125b binding site in the 3′ untranslated region (3′UTR) of bone morphogenetic protein membrane receptor type IB gene (BMPR1B) has been found to contribute to cancer susceptibility. To investigate whether it plays an important role in the development of prostate cancer in southern Chinese Han population, we performed a case–control study. 247 prostate cancer and 278 control subjects were included in the cancer association study and dual-luciferase reporter assay was used to test the binding ability of miR-125b to BMPR1B-C or -T vectors. The effect of CT/TT genotype on prostate cancer risk was found to be significant for localized disease (OR = 1.60, 95% CI = 1.01–2.53, P = 0.044) and among subgroups of aged >70 years (OR = 1.90, 95% CI = 1.15–3.15, P = 0.015) compared with CC genotype. Moreover, C-allele gave a reduced luciferase activity relative to T-allele in dual-luciferase reporter assay. Our findings show that rs1434536 in the 3′UTR of BMPR1B gene affects the binding ability of miR-125b to BMPR1B mRNA and contributes to the genetic predisposition to localized prostate cancer and patients aged >70 years.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
MicroRNAs (miRNAs) are a recently discovered class of small nocoding RNAs that regulate gene expression by binding to mRNA sequences and repressing target-gene expression post-transcriptionally, either by inhibiting translation or promoting RNA degradation [1, 2] and play fundamental roles in cell growth, apoptosis, hematopoietic lineage differentiation, and differentiation [3, 4]. There is emerging evidence that miRNAs are involved in cancer pathogenesis. Moreover, genomic variation within miRNA target sites may therefore be important sources for genetic differences in human cancer risk [5–7].
Prostate cancer is one of the most common cancers in men and is the second leading cause of male cancer death in the United States [8]. There were differential expression of miR-125b in androgen-dependent and independent prostate cancer cells as well as in benign and malignant prostate tissues [9], suggesting that miR-125b may contribute to the formation of prostate cancer. Recently, a C>T polymorphism (rs1434536) in a miR-125b binding site in the 3′ untranslated region of bone morphogenetic protein membrane receptor type IB gene (BMPR1B), which results in an reduced binding ability of miR-125b to BMPR1B-T allele, has been found to contribute to breast cancer susceptibility [10]. BMPR1B binds bone morphogenetic proteins (BMP) and all of them are multifunctional signaling molecules that belong to the transforming growth factor β (TGF-β) superfamily and were originally isolated as factors that induced bone and cartilage formation [11]. Consequent work has demonstrated that normal BMP/receptors are critical during mammalian development, cellular chemotaxis, and cellular differentiation [12–14]. Moreover, BMP/receptors were also detected in the cancerous prostate tissues and in the prostate cancer cell lines [15], suggesting that BMP/receptors may be involved in the osteogenic metastasis of the prostate cancer.
To date, there is no data about the association between BMPR1B polymorphism and prostate cancer risk. Hence, we explored the role of this polymorphism in prostate cancer in southern Chinese Han population and further examined the miR-125b binding ability on BMPR1B 3′UTR containing either C or T alleles.
Materials and methods
Study population
Two hundred and forty-seven prostate cancer patients were diagnosed between December 2006 and July 2009, and were pathologically proven to have prostate adenocarcinoma after biopsy in the First Affiliated Hospital of Nanjing Medical University (Jiangsu Province Hospital) in Nanjing, China. All the patients were southern Chinese Han population. The control group (n = 278) was age-matched, and the subjects were healthy checkup examinees without cancer history and were collected in the same period. Controls were excluded if they had an abnormal prostate specific antigen (PSA) level, or abnormal digital rectal examination (DRE).
After informed consent was obtained, 2 ml peripheral blood sample was collected and each subject was asked to finish a questionnaire including age, race, tobacco use, alcohol use, family history of cancer and so on. In present research, smoking more than five cigarettes per day for more than 5 years was defined as smoking. Pack-years of smoking (cigarettes per day ÷ 20) × (years with smoked) were calculated to indicate the cumulative smoking dose. Drinking habit was defined as drinking at least three times per week and lasting more than 10 years. Family history of cancer was defined as cancer in first-degree relatives (parents, siblings, or children). Disease stage was determined by pathologic findings, pelvic computed tomography, magnetic resonance image and radio-nucleotide bone scans. The tumor stage was determined using TNM classification and graded according to WHO guidelines. Pathologic grade was recorded as the Gleason score.
Genotyping
Genomic DNA was extracted from whole blood by standard proteinase K digestion and the phenol/chloroform method [16]. Genotyping was carried out using TaqMan SNP Genotyping Assays (Applied Biosystems). Reaction volumes of 10 μl, including 20 ng of DNA, TaqMan Universal Master Mix, and the SNP assay mix. PCR reactions were done on a PTC 200 Thermal Cycler (Bio-Rad inc.) with the following protocol: 2 min at 50°C, and 10 min at 95°C, and then 40 cycles of 15 s at 95°C and 1 min at 60°C. The SNP assays were analyzed on a 7300HT and alleles were called using Sequence Detection System version 2.3.
Dual luciferase reporter assays
We obtained DU145 and PC3 cell lines from Shanghai Cell Bank, Chinese Academy of Sciences and cultivated it in DMEM (Gibco), supplemented with 10% fetal bovine serum (Hangzhou Sijiqing Biological Engineering Materials Co., Ltd). Transfections were performed with lipofectamine 2000 (Invitrogen). Luciferase reporter vectors (psiCHECK-2) containing C or the T alleles at rs1434536 in the 3′-UTR of BMPR1B gene were granted by Dr. Garrett P. Larson. Du145 and PC3 cells were transfected with vectors with C or T allele. And 24 h later, they were transfected with miR-125b mimics or NC (negative control) (Shanghai GenePharma Co., Ltd). The sequence of miR-125b mimics was 5′-UCCCUGAGACCCUAACUUGUGA-3′ and that of NC was 5′-UUCUCCGAACGUGUCACGUTT-3′. The cells were collected 48 h after the second transfection, and cell lyases were prepared according to the manufacture’s instruction. Luciferase activity was measured with a Dual-Luciferase Reporter Assay System (Beyotime Co., Ltd) and normalized against the activity of the Renilla luciferase gene.
Statistical analysis
Hardy–Weinberg equilibrium was assessed using goodness-of-fit chi-square test. Distribution of demographic characteristics and substance genotypes were assessed by calculating the OR and 95% CI. Group means were compared by Student’s t-test. A P-value <0.05 was considered statistically significant, and all statistical tests were two sided. All statistical analyses were done using the Statistical Package of the Social Sciences software version 16.0 (SPSS, Inc.).
Results
Characteristics of the study population
The demographic characteristics of prostate cancer cases and healthy controls are shown in Table 1. Baseline characteristics were similar between cases and controls, except that there were more subjects who had large body mass index (BMI >23 kg/m2) among the cases than among the controls (66.80 vs. 57.91%, P = 0.036), and the frequency of relatives with cancer from the case group was higher, compared to nonrelatives (25.10 vs. 14.75%, P = 0.003).
Genotype distributions
The distribution of rs1434536 in the control group was 37.41% for CC homozygote, 46.40% for CT heterozygote, and 16.19% for TT homozygote, which did not appear to differ from that of the HapMap CHB population. The genotype distributions in the controls fit the Hardy–Weinberg equilibrium (χ2 = 0.22, P = 0.639).
As shown in Table 2, the TT genotype in the patients was more frequent than that in the controls (17.81 vs. 16.19%), although no statistically significance was found. A similar trend was also observed in the dominant genetic model. The association between the genotype and the risk of prostate cancer was further analyzed using multivariate unconditional logistic regression, with adjustment for potential covariates (age, BMI, cigarette smoking, alcohol drinking, family history of cancers). The CT/TT genotype was found tender to have a risk effect on prostate cancer, however, the result did not reach statistically significance (OR = 1.33, 95% CI = 0.92–1.92, P = 0.161).
Stratified analysis
Supplemental Table 1 lists the association between genotypes and prostate cancer risk stratified by disease stage, Gleason score and serum PSA level. The effect of rs1434536 CT/TT genotype on prostate cancer risk was found to be significant for localized disease (OR = 1.60, 95% CI = 1.01–2.53, P = 0.044 for CT/TT vs. CC) but not for advanced prostate cancer (Fig. 1). Moreover, no significant association between the SNP and the risk for prostate cancer was found when stratifying the case group by Gleason score and serum PSA level. Interestingly, as shown in Supplemental Table 2 and Fig. 2, the increased risk was more pronounced among subgroups of aged >70 years (OR = 1.90, 95% CI = 1.15–3.15, P = 0.015) when comparing the CT/TT genotype with the CC genotype, but not in subgroups of aged ≤70 years (OR = 0.89, 95% CI = 0.51–1.54). In addition, there was no significant association in the stratified analysis by BMI, status of cigarette smoking and alcohol drinking and family history of cancers.
Dual-luciferase reporter assay
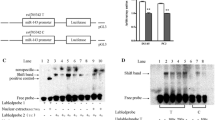
Vectors containing either C or T alleles and miR-125b or NC mimics transiently transfected into DU145 and PC3 cell lines and Renilla luciferase activity was measured. When transfected with miR-125b, the C-allele gave a reduced luciferase activity relative to the T allele (Fig. 3, P < 0.05), which suggested a direct functional effect of the SNP. However, when we transfected the two vectors with NC, no significant result was found between the two alleles.
Luciferase reporter assays to measure C to T allele differences at rs1434536. Du145 and PC3 cells were transiently transfected with C or T bearing reporter vectors, which is predicted to influence the recognition of the miR-125b seed sequence in the BMPR1B 3′-UTR and miR-125b mimics. Renilla luciferase activity was measured and normalized to Firefly luciferase. Results are shown as percentage relative to luciferase activity. Data represent the mean ± SEM of at least three independent experiments. *,**P < 0.05
Discussion
To date, this is the first study on BMPR1B polymorphism to prostate cancer susceptivity. In present study, we performed a case–control study with 247 prostate cancer cases and 278 healthy controls in southern Chinese Han population and rs1434536 CT/TT genotype was prominent in the prostate cancer group and revealed a higher risk of developing prostate cancer than CC genotype in individuals aged over 70 years old or with localized disease.
BMP/receptors belong to the TGF-Beta superfamily and are vital bone inductive factors, which also play important roles during embryonic development and the postnatal homeostasis of various organs and tissues, by controlling cellular differentiation, proliferation and apoptosis[17, 18]. Mutations in BMPR1B are rare, and few articles focused on polymorphisms of BMPR1B. Recently, a C>T polymorphism (rs1434536) in the 3′ untranslated region of BMPR1B has been found to contribute to breast cancer susceptibility [10]. A competition between the free energy gained by miRNA binding and the energetic cost of displacing existing RNA secondary structure at the target site was summarized by PITA [19]. And inputting the 200 nucleotides centered on rs1434536C/T alleles to PITA gave dG values of −0.53 and 3.09. As smaller values indicate stronger miRNA binding, it suggested that replacing C allele by T allele reduced binding ability of miR-125b to BMPR1B mRNA. It was confirmed by our further dual-luciferase reporter assay. The C-allele gave a reduced luciferase activity relative to the T allele, which suggested a direct functional effect of the SNP.
Rs1434536 CT/TT genotype was found to be associated with localized disease (OR = 1.60, 95% CI = 1.01–2.53, P = 0.044) but not advanced prostate cancer in our population. Although a correlation has been found between expression of BMPR1B and clinical severity of prostate cancer [15], how rs1434536 polymorphism affects the clinical features of prostate cancer remains unknown. Whether rs1434536 polymorphism had phenotypic effects on prostate cancer deserves further investigation. Age might be a risk factor for prostate cancer. We found the OR for rs1434536 CT/TT genotype was 1.90 (95% CI: 1.15–3.15, P = 0.015) among patients aged >70 years, which suggested that rs1434536 was a more appropriate predictor for prostate cancer in elder people since prostate cancer had an age-related increased incidence.
It was known to all that larger BMI, drinking, smoking and familiar cancer history were all considered as potential risk for various cancers. In present study, we found that more subjects whose BMI was over 23 kg/m2 among the cases than among the controls, and the frequency of relatives with cancer from the case group was higher, compared to nonrelatives. Unfortunately, we failed to find any evidence for interactions between the polymorphism and these factors. This observed phenomenon might indicate that these cancer patients without the established risk factors may have some other unknown exposures, or have other high level of genetic susceptibilities, or have other unknown risk factors that are linked to the putative risk genotypes. Another possibility is that the results are purely chance as the numbers in the subgroups compared were relatively small. Further, larger studies are wanted to confirm the results.
Conclusions
Our findings uncover that the C>T polymorphism (rs1434536) in the 3′ untranslated region of BMPR1B gene affects the binding ability of miR-125b to BMPR1B mRNA, contributes to the genetic predisposition to prostate cancer, and plays a role in the tumorigenesis. As GWAS have identified only common SNPs as genetic risk factors, it is likely that many rare alleles present within motifs for miRNAs remain to be identified. Since the number of the cases and controls are relatively small, larger studies are needed to confirm this relationship with different ethnic groups and more detailed environmental exposure data.
References
Schmittgen TD, Jiang J, Liu Q, Yang L (2004) A high-throughput method to monitor the expression of microRNA precursors. Nucleic Acids Res 32:e43
Wu W, Sun M, Zou GM, Chen J (2007) MicroRNA and cancer: current status and prospective. Int J Cancer 120:953–960
Ambros V (2004) The functions of animal microRNAs. Nature 431:350–355
Kloosterman WP, Plasterk RH (2006) The diverse functions of microRNAs in animal development and disease. Dev Cell 11:441–450
Landi D, Gemignani F, Naccarati A, Pardini B, Vodicka P, Vodickova L, Novotny J, Forsti A, Hemminki K, Canzian F, Landi S (2008) Polymorphisms within micro-RNA-binding sites and risk of sporadic colorectal cancer. Carcinogenesis 29:579–584
Yu Z, Li Z, Jolicoeur N, Zhang L, Fortin Y, Wang E, Wu M, Shen SH (2007) Aberrant allele frequencies of the SNPs located in microRNA target sites are potentially associated with human cancers. Nucleic Acids Res 35:4535–4541
George GP, Gangwar R, Mandal RK, Sankhwar SN, Mittal RD (2011) Genetic variation in microRNA genes and prostate cancer risk in North Indian population. Mol Biol Rep 38:1609–1615
Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ (2009) Cancer statistics 2009. CA Cancer J Clin 59:225–249
Shi XB, Xue L, Yang J, Ma AH, Zhao J, Xu M, Tepper CG, Evans CP, Kung HJ, deVere White RW (2007) An androgen-regulated miRNA suppresses Bak1 expression and induces androgen-independent growth of prostate cancer cells. Proc Natl Acad Sci USA 104:19983–19988
Saetrom P, Biesinger J, SM Li, Smith D, Thomas LF, Majzoub K, Rivas GE, Alluin J, Rossi JJ, Krontiris TG, Weitzel J, Daly MB, Benson AB, Kirkwood JM, O’Dwyer PJ, Sutphen R, Stewart JA, Johnson D, Larson GP (2009) A risk variant in an miR-125b binding site in BMPR1B is associated with breast cancer pathogenesis. Cancer Res 69:7459–7465
Wozney JM, Rosen V, Celeste AJ, Mitsock LM, Whitters MJ, Kriz RW, Hewick RM, Wang EA (1988) Novel regulators of bone formation: molecular clones and activities. Science 242:1528–1534
Hogan BL (1996) Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev 10:1580–1594
LI M, Eriksen EF, Bunger C (1996) Bone morphogenetic protein-2 but not bone morphogenetic protein-4 and -6 stimulates chemotactic migration of human osteoblasts, human marrow osteoblasts, and U2-OS cells. Bone 18:53–57
Paralkar VM, Weeks BS, Yu YM, Kleinman HK, Reddi AH (1992) Recombinant human bone morphogenetic protein 2B stimulates PC12 cell differentiation: potentiation and binding to type IV collagen. J Cell Biol 119:1721–1728
Kim IY, Lee DH, Ahn HJ, Tokunaga H, Song W, Devereaux LM, Jin D, Sampath TK, Morton RA (2000) Expression of bone morphogenetic protein receptors type-IA, -IB and -II correlates with tumor grade in human prostate cancer tissues. Cancer Res 60:2840–2844
Lee TH, Sakahara NS, Fiebig EW, Hirschkorn DF, Johnson DK, Busch MP (1998) Quantitation of white cell subpopulations by polymerase chain reaction using frozen whole-blood samples. Viral activation transfusion study. Transfusion 38:262–270
Ye L, Lewis-Russell JM, Kyanaston HG, Jiang WG (2007) Bone morphogenetic proteins and their receptor signaling in prostate cancer. Histol Histopathol 22:1129–1147
Murakami M, Matsuzaki F, Funaba M (2009) Regulation of melanin synthesis by the TGF-beta family in B16 melanoma cells. Mol Biol Rep 36:1247–1250
Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E (2007) The role of site accessibility in microRNA target recognition. Nat Genet 39:1278–1284
Acknowledgments
This study was supported by the foundation of medical key department of Jiangsu Province—department of general surgery of Jiangsu Province Hospital and the foundation of medical key department of Jiangsu Province—department of urology of Jiangsu Province Hospital (XK17 200904) and Natural Science Foundation of Jiangsu Province (No. SBK201021952). This publication is also part of my research work at Karolinska Institutet, thanks to a Swedish Institute scholarship.
Conflict of interest
The authors declare that they have no conflict of interest related to the publication of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Ninghan Feng, Bin Xu, Jun Tao and Pengchao Li contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Feng, N., Xu, B., Tao, J. et al. A miR-125b binding site polymorphism in bone morphogenetic protein membrane receptor type IB gene and prostate cancer risk in China. Mol Biol Rep 39, 369–373 (2012). https://doi.org/10.1007/s11033-011-0747-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-0747-9