Abstract
MicroRNAs (miRNAs) are small non-coding RNA molecules that function as negative regulators of gene expression. Common genetic variants (single nucleotide polymorphisms, SNPs) in miRNA genes may alter their expression or maturation resulting in varied functional consequences. Until now, several studies had evaluated the association between the polymorphisms in the hsa-miR-196a2 rs11614913 and cancer risk in diverse populations and in multiple types of cancer, with contradictory outcomes. Therefore, here we performed a meta-analysis to address the association between this polymorphism and cancer risk. A total of nine studies involving 6,540 cases and 7,562 controls were retrieved based on PubMed. Our analysis demonstrated that hsa-miR-196a2 rs11614913 CC genotype significantly increased the cancer risk in homozygote comparison model compared to TT genotype (OR = 1.18; 95% CI, 1.01–1.68). Moreover, significant association of this polymorphism with breast cancer was found based on homozygote comparison model (OR = 1.30; 95% CI, 1.01–1.26) and dominant model (OR = 1.11; 95% CI, 1.01–1.23). In addition, hsa-miR-196a2 rs11614913 CC genotype was significantly associated with cancer risk in Chinese and Indian (OR = 1.21; 95% CI, 1.05–1.40), but not in Caucasians (OR = 1.03; 95% CI, 0.89–1.19). Taken together, our results indicate that the polymorphism of hsa-miR-196a2 rs11614913 is associated with cancer susceptibility, especially with breast cancer and in Chinese and Indian populations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
MicroRNAs (miRNAs) are small single-stranded nonprotein-coding RNAs of about 22 nucleotides and play key roles in a broad range of physiologic and pathologic processes [1, 2]. These miRNAs regulate the expression of roughly 10–30% of all human genes through post-transcriptional mechanisms pairing to complementary sequences in the 3′ untranslated region (3′ UTR) of target mRNAs, leading to mRNA degradation or translational repression [3]. Emerging evidence has suggested the involvement of this novel class of gene regulators in cancer-related processes. It was shown that miRNA expression profiles could be used to classify human cancer [4], and a global repression of miRNA maturation induced cellular transformation and tumorigenesis [5]. The loss and gain of function of specific miRNAs were also thought to be key events in diverse cancers [6].
Although the precise processes controlling miRNAs expression and maturation are largely unknown, several mechanisms have been proposed including genetic and epigenetic alterations [7, 8]. The most common genetic variation, single nucleotide polymorphisms (SNPs), is known to exist in miRNA genes, which can lead to variations in the quantity of miRNAs resulting in diverse functional consequences.
Recent data have shown that SNPs located in the pre-miRNA, such as hsamiR-196a2 rs11614913 polymorphism may influence the expression of mature miRNA [8–11]. Furthermore, the association between hsa-miR-196a2 rs11614913 polymorphism and cancer risk has been analyzed in several studies although the results are controversial [12–19]. Because the relatively small sample size in a single study might have low power to detect the effect of the polymorphisms on cancer risk, here we reported our meta-analysis that may improve our evaluation of the association of miRNA-196a2 polymorphism with cancer risk.
Materials and methods
Eligible studies and data extraction
To identify all previous published studies that examined the association of hsa-miR-196a2 rs11614913 polymorphism with cancer, PubMed database (from July 2008 to June 2010) was searched with the following keywords and subject terms: ‘‘microRNA’’ and ‘‘cancer’’ and “polymorphism’’. The following criteria were set to choose the studies included in the current meta-analysis: (1) evaluation of the hsa-miR-196a2 rs11614913 polymorphism and cancer risk; (2) a case–control design; (3) malignant tumors were histologically confirmed; (4) sufficient published data for calculating odds ratios (ORs) with their 95% confidence intervals (95% CIs).
The following data were abstracted independently in duplicate by two investigators using a standard protocol and data-collection form according to the inclusion criteria listed above: First author’s surname, publication date, ethnicity, tumor type, genotyping methods, characteristics of cases and controls, and P value for Hardy–Weinberg equilibrium were described (Table 1). Breast cancer was performed to pool cancer-specific ORs and different racial descents were categorized as Asian and Caucasian. When more than one ethnicity was included in the studies and was able to separate, data were extracted separately for each ethnic group. Disagreement was resolved by the discussion between the two investigators.
Statistical analysis
The strength of association between hsa-miR-196a2 rs11614913 polymorphism and cancer risk was assessed by crude ORs with their 95% CIs. The pooled ORs were calculated for homozygote comparison (CC vs. TT), heterozygote comparison (CC vs. CT), dominant model (CC vs. CT + TT), and recessive model (CC + CT vs. TT), respectively. The statistical heterogeneity among the studies was checked by the chi-square-based Q-test [20].
Heterogeneity was considered significant for P < 0.10 because of the low power of the statistic. In the absence of between-study heterogeneity, the fixed-effects model using the Mantel–Haenszel method was used to estimate the summarized OR [21]; and Random-effects using the DerSimonian and Laird method was more appropriate when heterogeneity is present [22].
The significance of the pooled OR was determined by the Z test with P value <0.05 considered significant. For each genetic comparison, subgroup analysis according to different kinds of cancer was firstly performed to pool cancer-specific ORs for breast cancer since more independent studies examined this kind of cancer. Subgroup analysis according to ethnicity was also investigated to estimate ethnic-specific ORs for Asian and Caucasian.
Publication bias of literatures was assessed using Begg’s funnel plot, and the significance of the intercept was determined by the t test as suggested by Egger, and a P value <0.05 was considered significant [23]. Hardy–Weinberg equilibrium was tested by the chi-square test for goodness of fit using a web-based program (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl).
All statistical analyses were carried out with STATA software, version 10.0 (STATA Corp., College Station, TX). All P values were two-sided.
Results
Selection of studies
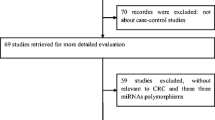
In total, ten case–control publications [12–19, 24, 25] were retrieved from Pubmed based on the searching criteria and eight met our inclusion criteria excluding two publications with no detailed genotyping information. In the study of Catucci et al., the genotype frequencies were presented separately according to German study and Italian study [17], and thus each of these studies was considered separately for meta-analysis. Therefore, a total of nine studies were included in the meta-analysis with 6,540 cases and 7,562 controls for hsa-miR-196a2 rs11614913 polymorphism, among which there were five studies of Asians [12, 13, 15, 16, 18], three studies of Caucasians [17, 19], and one study of Mixed population with no detailed ethnicity data [14].
Multiple genotyping methods had been employed in the studies included in our analysis: polymerase chain reaction-restriction fragment length polymorphism (PCR–RFLP), MassARRAY multiplex, Taqman SNP genotyping assay, and DNA sequencing. The studies of Hoffman et al., Hu et al. and Srivastava et al. mentioned quality control for the genotyping assay. Blood sample was used for genotyping in all the studies. The distribution of genotypes in the controls of all the studies was in agreement with HWE.
Main results
The main results of this meta-analysis were shown in Table 2. For hsa-miR-196a2 rs11614913 polymorphism, significantly increased cancer risk was observed in homozygote comparison (OR = 1.180; 95% CI (1.006–1.384), P = 0.041, p-heterogeneity = 0.021), and recessive model (OR = 1.137; 95% CI (1.045–1.237), P = 0.003, p-heterogeneity = 0.352) (Fig. 1).
a Overall meta-analyses for hsa-miR-196a2 rs11614913 polymorphism (CC vs. TT) in cancers under a random-effects model. b Overall meta-analyses for hsa-miR-196a2 rs11614913 polymorphism (CC + CT vs. TT) in cancers under a fixed-effects model. Each retrieved study was presented by the surname of the first author and the year of publication, and the corresponding OR and 95% CI were shown
Next we performed the meta-analysis under other genetic comparisons. Firstly, in different types of cancer, hsa-miR-196a2 rs11614913 polymorphism was significantly associated with an increased risk of breast cancer both in homozygote comparison (OR = 1.304, 95% CI (1.011–1.682), P = 0.002, p-heterogeneity = 0.028) and dominant model (OR = 1.114; 95% CI, 1.011–1.227, P = 0.029, p-heterogeneity = 0.21) (Fig. 2). Secondly, hsa-miR-196a2 rs11614913 polymorphism genotypes were significantly associated with cancer risk in some stratified population. In Asians including Chinese and Indian, in homozygote comparison model by comparing CC to TT the OR was 1.212 (95% CI (1. 051–1.395), P = 0.008, p-heterogeneity = 0.141), and in recessive model by comparing CC + CT to TT the OR was 1.156 (95% CI (1.035–1.291), P = 0.01, p-heterogeneity = 0.942). However, no significant association of hsa-miR-196a2 rs11614913 polymorphism with cancer risk was found in Caucasians (Table 2; Fig. 3).
a Meta-analyse for hsa-miR-196a2 rs11614913 polymorphism (CC vs. TT) in breast cancer under a random-effects model. b Meta-analyse for hsa-miR-196a2 rs11614913 polymorphism (CC vs. CT + TT) in breast cancer under a fixed-effects model. Each retrieved study was presented by the surname of the first author and the year of publication, and the corresponding OR and 95% CI were shown
Begg’s funnel plot was performed to evaluate the potential publication bias of literatures, and no evidence of publication bias was observed in any comparison model (P < 0.05) (Fig. 4). Finally, sensitivity analysis was carried out by investigating the influence of each study on the overall OR, and the result showed that no individual study affected the overall OR dominantly, since the omission of any single study made no substantial difference (Fig. 5).
The influence of individual studies on the summary OR. The middle vertical axis indicates the overall OR and the two vertical axes indicate its 95% CI. Every hollow round indicates the pooled OR when the left study was omitted in this meta-analysis. The two ends of every broken line represent the 95% CI
Discussion
SNPs are the most common sequence variation in the human genome affecting coding sequences, splicing, and even noncoding sequences such as miRNAs. Notably, a role for SNPs in cancer predisposition and particularly in cancer prognosis has been proposed [26]. Hoffman and colleagues found that the sequence variant in hsa-miR-196a2 (rs11614913) located in the 3p mature miRNA region may affect pre-miRNA maturation of 5p and 3p miRNAs as well as the expression and function of these miRNAs in breast cancer [14]. Additional studies showed that hsa-miR-196a2 rs11614913 polymorphism occur in different kinds of cancer with potential prognosis value [12–19, 24, 25, 27].
Given the controversial results in previous studies, we performed this meta-analysis to address the association of this sequence variant with cancer risk and quantify the potential between-study heterogeneity. Our results demonstrate that individuals carrying CC genotype of hsa-miR-196a2 rs11614913 polymorphism had increased cancer risk, indicating that this common genetic variant may play crucial roles in cancer development.
Due to the limited number of studies included in this analysis, we performed sub-analysis by cancer types just for breast cancer. The pooled ORs of 4 studies suggested a significant association between homozygote comparison as well as dominant model and breast cancer, consistent with the results previously reported [28]. Interestingly, Hoffman et al. reported that the expression of some genes indicating poor clinical outcome in breast tumors was significantly altered following the introduction of miR-196a-C but was unchanged in the miR-196a-T cells [14]. Taken together, these data strongly suggest that hsa-miR-196a2 rs11614913 polymorphism has the potential relevance to breast tumor progression.
After stratification by ethnicity, the allelic frequency of hsa-miR-196a2 rs11614913 polymorphism was different in Asians and Caucasians. CC genotype was significantly associated with cancer risk in Chinese and Indian but not in Caucasians, suggesting potentially different mechanisms underlying tumorigenesis in different populations.
Some limitations of this meta-analysis should be considered. First, the number of studies included in the meta-analysis was too small to perform subgroup analysis including every type of cancer. Second, a more precise analysis which would allow for the adjustment by other covariates such as age, sex, family history, environmental factors and lifestyle should be conducted if individual data were available. Third, so far there was no study of other populations except Asian and Caucasians. Therefore, further studies regarding the relationships among the hsa-miR-196a2 rs11614913 genetic variations, miR-196a2 levels, the risk of cancer, and the factors mentioned above will be urgently needed.
In conclusion, our meta-analysis suggested that hsa-miR-196a2 rs11614913 polymorphism is associated with an increased cancer risk. Further stratification to ethnics (Caucasians and Asians) and cancer types (breast cancer) in different genetic background also identified the significant association of this polymorphism with cancer risk, especially in Asians and with breast cancer. Future well-designed case–control studies with larger sample size are of great value to confirm these findings.
References
Ambros V (2004) The functions of animal microRNAs. Nature 431(7006):350–355
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136(2):215–233
Duan R, Pak C, Jin P (2007) Single nucleotide polymorphism associated with mature miR-125a alters the processing of pri-miRNA. Hum Mol Genet 16(9):1124–1131
Kent OA, Mendell JT (2006) A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene 25(46):6188–6196
Lu J et al (2005) MicroRNA expression profiles classify human cancers. Nature 435(7043):834–838
Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T (2007) Impaired microRNA processing enhances cellular transformation, tumorigenesis. Nat Genet 39(5):673–677
Lujambio A et al (2007) Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res 67(4):1424–1429
Xu T et al (2008) A functional polymorphism in the miR-146a gene is associated with the risk for hepatocellular carcinoma. Carcinogenesis 29(11):2126–2131
Jazdzewski K et al (2008) Common SNP in pre-miR-146a decreases mature miR expression and predisposes to papillary thyroid carcinoma. Proc Natl Acad Sci USA 105(20):7269–7274
Shen J et al (2008) A functional polymorphism in the miR-146a gene, age of familial breast/ovarian cancer diagnosis. Carcinogenesis 29(10):1963–1966
Peng S et al (2010) Association of microRNA-196a-2 gene polymorphism with gastric cancer risk in a chinese population. Dig Dis Sci 55(8):2288–2293
Tian T et al (2009) A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol Biomarkers Prev 18(4):1183–1187
Hoffman AE et al (2009) microRNA miR-196a-2 and breast cancer: a genetic and epigenetic association study and functional analysis. Cancer Res 69(14):5970–5977
Hu Z et al (2009) Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum Mutat 30(1):79–84
Dou T et al (2010) A polymorphism of microRNA196a genome region was associated with decreased risk of glioma in Chinese population. J Cancer Res Clin Oncol 136(12):1853–1859
Catucci I et al (2010) Evaluation of SNPs in miR-146a, miR196a2 and miR-499 as low-penetrance alleles in German and Italian familial breast cancer cases. Hum Mutat 31(1):E1052-7
Srivastava K, Srivastava A, Mittal B (2010) Common genetic variants in pre-microRNAs and risk of gallbladder cancer in North Indian population. J Hum Genet 55(8):495–499
Liu Z et al (2010) Genetic variants in selected pre-microRNA genes and the risk of squamous cell carcinoma of the head and neck. Cancer 116(20):4753–4760
Higgins JP, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21(11):1539–1558
Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22(4):719–748
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7(3):177–188
Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315(7109):629–634
Hu Z et al (2008) Genetic variants of miRNA sequences, non-small cell lung cancer survival. J Clin Invest 118(7):2600–2608
Christensen BC et al (2010) Mature miRNA sequence polymorphism in MIR196A2 is associated with risk and prognosis of head and neck cancer. Clin Cancer Res 16(14):3713–3720
O’Mara TA, Clements JA, Spurdle AB (2009) The use of predictive or prognostic genetic biomarkers in endometrial, other hormone-related cancers: justification for extensive candidate gene single nucleotide polymorphism studies of the matrix metalloproteinase family, their inhibitors. Cancer Epidemiol Biomarkers Prev 18(9):2352–2365
Landgraf P et al (2007) A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129(7):1401–1414
Gao LB et al (2011) The association between two polymorphisms in pre-miRNAs and breast cancer risk: a meta-analysis. Breast Cancer Res Treat 125(2):571–574
Acknowledgments
This work was supported by grants from the Shanghai Science and Technology Development Fund (No. 05DJ14010), the Major Basic Research Program of Shanghai (No. 07DZ19505), and the National 973 Basic Research Program of China (No. 2008CB517403).
Conflict of interest
None of the authors has any potential conflict of interest related to this manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Wang, F., Ma, YL., Zhang, P. et al. A genetic variant in microRNA-196a2 is associated with increased cancer risk: a meta-analysis. Mol Biol Rep 39, 269–275 (2012). https://doi.org/10.1007/s11033-011-0735-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-0735-0