Abstract
The aim of this study was to determine whether the interleukin-10 (IL-10) polymorphisms confer susceptibility to rheumatoid arthritis (RA). A meta-analysis was conducted on the associations between the IL-10 −1082 G/A, −592 C/A, −892 C/T and IL-10.R polymorphisms and RA using; (1) allele contrast, (2) the recessive model, (3) the dominant model, and (4) the additive model. A total of 16 studies (19 comparisons) involving 2647 RA patients and 3383 controls were considered in the meta-analysis. Meta-analysis of the IL-10 −1082 G/A polymorphism showed no association with RA in the study subjects, or in European or Asian subjects. However, meta-analysis of the −1082 G allele in 4 studies in Hardy–Weinberg equilibrium showed a significant association with RA (OR = 1.217, 95% CI = 1.027–1.442, P = 0.0236). In contrast, meta-analysis of the C allele, the CC genotype, and of the CC versus the AA genotype of the IL-10 −592 C/A polymorphism showed significant associations with RA. The overall ORs of the associations between the C allele and RA were 0.684 and 0.758 (95% CI = 0.494–0.946, P = 0.022; 95% CI = 0.475–1.210, P = 0.045) in all study subjects and Asians. Meta-analysis of the CC + CT versus TT genotype and of the CC versus TT genotype of the IL-10 −892 C/T polymorphism revealed significant associations with RA. The overall OR of the association between the C allele carrier and RA was 0.552 (95% CI = 0.375–0.812, P = 0.003). No association was found between the IL10.R2 alleles and RA. This meta-analysis suggests that the IL-10 −592 C/A polymorphism confers susceptibility to RA in Asians and that the IL-10 −1082 G/A and −892 C/T polymorphisms are associated with RA susceptibility. These findings suggest the IL-10 genes confer susceptibility to RA.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rheumatoid arthritis (RA) is a chronic inflammatory disease that predominantly involves synovial joints and affects up to 1% of adults worldwide [1]. Although the etiology of RA remains unknown, a genetic component has been established by twin and family studies, in which RA liability was estimated to be as high as 60%. Human leukocyte antigen (HLA) class II molecules have been shown to be strongly associated with RA, but family studies suggest that this association accounts for only one-third of genetic susceptibility and that non-HLA genes are also involved [2].
Interleukin-10 (IL-10) is a multifunctional cytokine that has anti-inflammatory properties due its ability to downregulate antigen presentation and macrophage activation [3]. IL-10 plays an important role in B cell activation and autoantibody production as a survival and differentiation factor, and it also acts as an inhibitory factor during the production of T helper 1 (Th1) cytokines [4]. The IL-10 gene maps to 1q31-32 and exhibits polymorphisms in its promoter region that appears to be correlated with variations in transcription. Three of several polymorphisms of IL-10 have been studied in some detail, namely, −1082 G to A (rs1800896), −592 C to A (rs1800872), and −892 C to T (rs1800871), and all three are located at putative regulatory regions in IL-10 promoter [5]. The −1082 G/A polymorphism lies within a putative Ets transcription factor binding site, while −592 C/A is located within a putative STAT-3 binding site and negative regulatory region, and -892 C/T lies within a putative positive regulatory region [6, 7]. Furthermore, the IL-10.R microsatellite polymorphism, situated 4 kb removed from the transcription initiation site in the 5′ direction, is also of interest, since haplotypes containing the IL-10.R.2 allele are associated with higher levels of IL-10 secretion than those containing IL-10.R3 [8]. Thus, polymorphisms at these sites may alter the binding sites of transcription factors that may affect IL-10 production. IL-10 is considered an attractive candidate gene based on its chromosomal location and functional relevance. A number of studies have examined the association between IL-10 polymorphisms and RA, but reported results are contradictory [9–24], possibly because of the low statistical powers of individual studies. Therefore, in order to overcome the limitations of individual studies, resolve inconsistencies, and reduce the likelihood that random errors are responsible for false-positive or false-negative associations [25–27], we turned to meta-analysis. In the present study, we used meta-analysis to explore whether the IL-10 −1082 G/A, −592 C/A, and IL-10.R polymorphisms contribute to RA susceptibility.
Methods
Identification of eligible studies and data extraction
A search was performed for studies that examined associations between IL-10 polymorphisms and RA. The literature was searched using the MEDLINE citation database to identify available articles in which IL-10 polymorphisms were analyzed in RA patients. Combinations of keywords, such as, “interleukin-10,” “IL-10,” “polymorphism,” “rheumatoid arthritis,” and “RA” were entered as Medical Subject Heading (MeSH) components and as text words. References in identified studies were also investigated to identify additional studies not indexed by MEDLINE. Genetic association studies that determined the distributions of IL-10 −1082 G/A, −592 C/A, −892 C/T, and the IL-10.R polymorphisms in RA and in normal controls were included. The following information was extracted from each study: author, year of publication, ethnicity of the study population, demographics, number of cases and controls, and the genotype and allele frequencies of each of the IL-10 −1082 G/A, −592 C/A, −892 C/T and IL-10.R polymorphisms.
Evaluation of publication bias
Funnel plots are often used to detect publication bias. However, due to the limitations of funnel plotting, which requires a range of studies of varying sizes involving subjective judgments, we evaluated publication bias using Egger’s linear regression test [28], which measures funnel plot asymmetry using a natural logarithm scale of odds ratios (ORs).
Evaluations of statistical associations
Allele frequencies at the IL-10 polymorphisms were determined by the allele counting method. The Chi-square test was used to determine if observed frequencies of genotypes in controls conformed to Hardy–Weinberg (H–W) expectations.
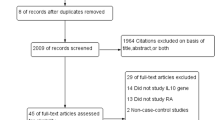
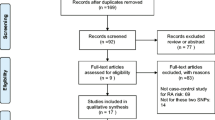
Meta-analyses was performed using; (1) allelic contrast and (2) recessive, (3) dominant, and (4) additive models. Point estimates of risks, ORs, and 95% confidence intervals (CI) were estimated for each study. Cochran’s Q-statistic was also used to assess within- and between-study variations and heterogeneities. This heterogeneity test assesses the null hypothesis that all studies evaluated the same effect. The effect of heterogeneity was quantified using I 2, which ranges from 0 to 100% and represents the proportion of between-study variability attributable to heterogeneity rather than chance [29]. I 2 values of 25, 50, and 75% were nominally considered low, moderate, and high estimates. The fixed effects model assumes that a genetic factor has a similar effect on RA susceptibility across all studies investigated, and that observed variations among studies are caused by chance alone [30]. On the other hand, the random effects model assumes that different studies show substantial diversity and assesses both within-study sampling errors and between-study variances [31]. When study groups are homogeneous, the two models are similar, but if this is not the case the random effects model usually provides wider CIs than the fixed effects model. The random effects model is best used in the presence of significant between study heterogeneity [31]. Statistical manipulations were performed using a Comprehensive Meta-Analysis computer program (Biosta, Englewood, NJ, USA) (Fig. 1).
Results
Studies included in the meta-analysis
Twenty-two relevant studies, which investigated the relation between an IL-10 polymorphism and RA, were identified using MEDLINE and by manual searching. Six studies were excluded due to; another IL-10 polymorphism, data duplication, or the lack of suitable controls. Thus, sixteen studies met our inclusion criteria [9–24]. One of these studies contained data on three different groups [23], and another data on two different groups [21], and these groups were analyzed independently. Therefore, a total of 19 separate comparisons were considered in this meta-analysis, which in total involved 2,647 RA patients and 3,383 controls, and eleven European, three Asian, two black, one Macedonian, one Turkish, and one Colombian population (Table 1). However, because populations were inadequate in the Macedonian, Turkish, and Colombian studies, ethnicity-specific meta-analysis was conducted on European, Asian, and Black populations. Details of the IL-10 polymorphisms studies are summarized in Table 1.
Meta-analysis of the IL-10 −1082 G/A, −592 C/A, −892 C/T and IL-10.R polymorphisms and RA susceptibility
A summary of meta-analyses findings concerning associations between IL-10 polymorphisms and RA is provided in Table 2. Meta-analysis of the IL-10 −1082 G/A polymorphism in the 6,030 study subjects revealed no association between RA and the IL-10 −1082 G allele (OR = 1.033, 95% CI = 0.863–1.236, P = 0.723). Furthermore, stratification by ethnicity indicated no association between the IL-10 −1082 G allele and RA in Europeans or Asians (Table 2), and no association was found between RA and the IL-10 −1082 G/A polymorphism using recessive or dominant models or contrast of homozygotes. In terms of the IL-10 −1082 G/A polymorphism, no association was found with RA by meta-analyses using the allele contrast, recessive, dominant, or additive models in all study subjects, or in European or Asian populations (Table 2). However, meta-analysis of the four studies in H–W did produce a result for the relation between the IL-10 −1082 G/A polymorphism and RA (Table 2). Specifically, analysis of the −1082 G allele revealed a significant association with RA (OR = 1.217, 95% CI = 1.027–1.442, P = 0.0236).
Meta-analysis of the C allele, the CC genotype, and the CC vs. AA genotype of the IL-10 −592 C/A polymorphism showed significant association with RA (Table 2). The overall ORs of the associations between the C allele and RA were 0.684 and 0.758 (95% CI = 0.494–0.946, P = 0.022; 95% CI = 0.475–1.210, P = 0.045) in all study subjects and in Asians (Fig. 2), and the ORs of the CC vs. AA genotype showed the same pattern as that observed for the C allele of the IL-10 −592 C/A polymorphism. Meta-analysis of the CC vs.CA + AA genotype also showed a significant association between the IL-10 −592 C/A polymorphism and RA in Asians (OR = 0.574, 95% CI = 0.37–0.873, P = 0.010). Meta-analysis of the CC + CT versus TT genotype, and the CC versus TT genotype of the IL-10 −89 C/T polymorphism also showed significant associations with RA (Table 3). The overall OR of the association between the C allele carrier and RA was 0.552 (95% CI = 0.375–0.812, P = 0.003) (Fig. 2). It has previously been shown that the IL10.R2 allele is associated with high IL10 secretion [8]. We performed meta-analysis on the association between the R2 allele of IL10.R and RA, but no association was found (Table 3).
Heterogeneity and publication bias
Some heterogeneity was found in several meta-analyses of the IL-10 −1082 G/A, −592 C/A, and IL-10.R polymorphisms and in the meta-analysis of C vs. T alleles of the IL-10 −892 C/T polymorphism in Asians. However, Egger’s regression test showed no evidence of publication bias in this meta-analysis (Egger’s regression test P-values > 0.1). The distributions of genotypes in the normal control groups were not consistent with the H–W equilibrium in eight studies of the IL-10 −1082 G/A polymorphism [9, 11, 12, 14, 16, 17, 19, 22]. Deviation from H–W equilibrium among controls implies potential bias during control selection, or genotyping errors, but excluding these studies did affect our result for an association between the IL-10-1082 G/A polymorphism and RA (Table 2). All studies were in H–W equilibrium for the IL-10 −592 C/A and −892 C/T polymorphisms.
Discussion
Although the multifactorial nature of RA is well recognized, genetic factors are considered to be strong determinants of these diseases, and thus, researchers have been encouraged to search for the genes responsible. Il-10 is a potent anti-inflammatory cytokine that inhibits the synthesis of pro-inflammatory cytokines, and a potent up-regulator of B-cell production and differentiation [4]. IL-10 may modulate disease severity in RA. It has been reported to suppress joint swelling and deformation and cartilage necrosis in an animal model of RA, and to be upregulated in the serum and synovial fluid of RA patients [32]. IL-10 production is genetically determined and is controlled at the transcription level, probably via some regulatory sequences in its promoter region [33].
In this meta-analysis, we addressed the association between IL-10 polymorphisms and RA susceptibility. Data from published studies were combined to evaluate genetic associations between the most commonly studied polymorphisms of the IL-10 gene, namely, the −1082 G/A, −592 C/A, −892 C/T, and IL-10.R polymorphisms and RA. Meta-analysis of the IL-10 −1082 G/A polymorphisms showed no association with RA in all study subjects, or in European or Asian subjects. However, meta-analysis of the −1082 G allele in the four studies in Hardy–Weinberg equilibrium revealed a significant association with RA. In contrast, meta-analysis of the IL-10 −592 C/A polymorphism revealed a significant association with RA in Asians, in whom, C allele carriage may be a protective factor with an OR of 0.758 (95% CI = 1.158–1.949, P = 0.045). Furthermore, whereas meta-analysis of the IL-10 −892 C/T polymorphism revealed a significant association with RA, no association was found between the IL10.R2 alleles and RA. The associations between IL-10 polymorphisms and the risks of RA observed in this meta-analysis suggest that IL-10 could play a role in RA susceptibility.
However, our results should be interpreted with caution because of the limited number of studies included, which also restricted further subgroup analyses. In addition, the distributions of the −1082 G/A genotypes in the normal control groups did not meet the requirement for H–W equilibrium in most studies. Because a deviation from H–W equilibrium among controls implies potential bias during control selection or genotyping errors, we re-performed subgroup analysis for studies in H–W equilibrium, but only four studies were in H–W equilibrium for the −1082 G/A polymorphism. For these four studies, although meta-analysis showed significant associations between the −1082 G/A polymorphism and RA [13, 15, 18, 24], this association was based on the results of one study only [15]. Furthermore, the relative importances of the IL-10 polymorphisms during the development of RA may vary between ethnic groups, but we failed to perform ethnic specific meta-analysis on the −892 C/T polymorphism due to limited data.
Present study has some limitations that require consideration. (1) Heterogeneity and confounding factors may have distorted the meta-analysis. Furthermore, publication bias also may have affected the analysis, because studies that produced negative results may not have been published or may have been missed, and although we performed Egger’s regression test, we could not eliminate the possibility of bias. (2) This ethnicity-specific meta-analysis included data from European and Asian patients, and thus, our results are applicable to only these ethnic groups. (3) Haplotype analysis may have provided more information and would have been more powerful than single polymorphism analysis. Linkage disequilibrium was found for the −1082 G/A, −592 C/A, −892 C/T polymorphisms. Increased IL-10 secretion has been described for the common GCC haplotype and reduced IL-10 secretion for the least common ATA haplotypes. No meta-analysis of haplotypes was possible due to the inadequacy of haplotype data. (4) IL-10 polymorphisms may be associated with RA severity as well as susceptibility. However, the small amount of data available did not allow us to perform meta-analysis this association.
In conclusion, this meta-analysis suggests that the IL-10 −592 C/A polymorphism confers susceptibility to RA in Asian populations. Furthermore, associations were found between the IL-10 −1082 G/A and −892 C/T polymorphisms and susceptibility to RA. However, our results should be interpreted with caution due to small number of studies included, and thus, our inability to perform subgroup analysis by ethnicity. Larger scale studies in populations with different ethnicities are necessary to explore the roles played by these polymorphisms of the IL-10 gene in the pathogeneses of RA.
References
Harris ED Jr (1990) Rheumatoid arthritis. Pathophysiology and implications for therapy. N Engl J Med 322:1277–1289
Deighton CM, Walker DJ, Griffiths ID, Roberts DF (1989) The contribution of HLA to rheumatoid arthritis. Clin Genet 36:178–182
Moore KW, de Waal Malefyt R, Coffman RL, O’Garra A (2001) Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol 19:683–765
Taga K, Tosato G (1992) IL-10 inhibits human T cell proliferation and IL-2 production. J Immunol 148:1143–1148
Eskdale J, Wordsworth P, Bowman S, Field M, Gallagher G (1997) Association between polymorphisms at the human IL-10 locus and systemic lupus erythematosus. Tissue Antigens 49:635–639
Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV (1997) An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet 24:1–8
Guzowski D, Chandrasekaran A, Gawel C, Palma J, Koenig J, Wang XP, Dosik M, Kaplan M, Chu CC, Chavan S, Furie R, Albesiano E, Chiorazzi N, Goodwin L (2005) Analysis of single nucleotide polymorphisms in the promoter region of interleukin-10 by denaturing high-performance liquid chromatography. J Biomol Tech 16:154–166
Eskdale J, Gallagher G, Verweij CL, Keijsers V, Westendorp RG, Huizinga TW (1998) Interleukin 10 secretion in relation to human IL-10 locus haplotypes. Proc Natl Acad Sci USA 95:9465–9470
Paradowska-Gorycka A, Trefler J, Maciejewska-Stelmach J, Lacki JK (2010) Interleukin-10 gene promoter polymorphism in Polish rheumatoid arthritis patients. Int J Immunogenet 37:225–231
Ying B, Shi Y, Pan X, Song X, Huang Z, Niu Q, Cai B, Wang L (2011) Association of polymorphisms in the human IL-10 and IL-18 genes with rheumatoid arthritis. Mol Biol Rep 38:379–385
de Paz B, Alperi-Lopez M, Ballina-Garcia FJ, Prado C, Mozo L, Gutierrez C, Suarez A (2010) Interleukin 10 and tumor necrosis factor-alpha genotypes in rheumatoid arthritis—association with clinical response to glucocorticoids. J Rheumatol 37:503–511
Menegatti E, Davit A, Francica S, Berardi D, Rossi D, Baldovino S, Tovo PA, Sena LM, Roccatello D (2009) Genetic factors associated with rheumatoid arthritis and systemic vasculitis: evaluation of a panel of polymorphisms. Dis Markers 27:217–223
Gambhir D, Lawrence A, Aggarwal A, Misra R, Mandal SK, Naik S (2010) Association of tumor necrosis factor alpha and IL-10 promoter polymorphisms with rheumatoid arthritis in North Indian population. Rheumatol Int 30:1211–1217
Trajkov D, Mishevska-Perchinkova S, Karadzova-Stojanoska A, Petlichkovski A, Strezova A, Spiroski M (2009) Association of 22 cytokine gene polymorphisms with rheumatoid arthritis in population of ethnic Macedonians. Clin Rheumatol 28:1291–1300
Ates O, Hatemi G, Hamuryudan V, Topal-Sarikaya A (2008) Tumor necrosis factor-alpha and interleukin-10 gene promoter polymorphisms in Turkish rheumatoid arthritis patients. Clin Rheumatol 27:1243–1248
Hee CS, Gun SC, Naidu R, Gupta E, Somnath SD, Radhakrishnan AK (2007) Comparison of single nucleotide polymorphisms in the human interleukin-10 gene promoter between rheumatoid arthritis patients and normal subjects in Malaysia. Mod Rheumatol 17:429–435
Moreno OM, Gonzalez CI, Saaibi DL, Otero W, Badillo R, Martin J, Ramirez G (2007) Polymorphisms of IL-10 gene promoter and rheumatoid arthritis in a Colombian population. Biomedica 27:56–65
Pawlik A, Kurzawski M, Szklarz BG, Herczynska M, Drozdzik M (2005) Interleukin-10 promoter polymorphism in patients with rheumatoid arthritis. Clin Rheumatol 24:480–484
Padyukov L, Hytonen AM, Smolnikova M, Hahn-Zoric M, Nilsson N, Hanson LA, Tarkowski A, Klareskog L (2004) Polymorphism in promoter region of IL10 gene is associated with rheumatoid arthritis in women. J Rheumatol 31:422–425
Martinez A, Pascual M, Pascual-Salcedo D, Balsa A, Martin J, de la Concha EG (2003) Genetic polymorphisms in Spanish rheumatoid arthritis patients: an association and linkage study. ***Genes Immun 4:117–121
MacKay K, Milicic A, Lee D, Tikly M, Laval S, Shatford J, Wordsworth P (2003) Rheumatoid arthritis susceptibility and interleukin 10: a study of two ethnically diverse populations. Rheumatology (Oxford) 42:149–153
Cantagrel A, Navaux F, Loubet-Lescoulie P, Nourhashemi F, Enault G, Abbal M, Constantin A, Laroche M, Mazieres B (1999) Interleukin-1beta, interleukin-1 receptor antagonist, interleukin-4, and interleukin-10 gene polymorphisms: relationship to occurrence and severity of rheumatoid arthritis. Arthritis Rheum 42:1093–1100
Eskdale J, McNicholl J, Wordsworth P, Jonas B, Huizinga T, Field M, Gallagher G (1998) Interleukin-10 microsatellite polymorphisms and IL-10 locus alleles in rheumatoid arthritis susceptibility. Lancet 352:1282–1283
Coakley G, Mok CC, Hajeer AH, Ollier WE, Turner D, Sinnott PJ, Hutchinson IV, Panayi GS, Lanchbury JS (1998) Interleukin-10 promoter polymorphisms in rheumatoid arthritis and Felty’s syndrome. Br J Rheumatol 37:988–991
Lee YH, Rho YH, Choi SJ, Ji JD, Song GG, Nath SK, Harley JB (2007) The PTPN22 C1858T functional polymorphism and autoimmune diseases–a meta-analysis. Rheumatology (Oxford) 46:49–56
Nath SK, Harley JB, Lee YH (2005) Polymorphisms of complement receptor 1 and interleukin-10 genes and systemic lupus erythematosus: a meta-analysis. Hum Genet 118:225–234
Lee YH, Witte T, Momot T, Schmidt RE, Kaufman KM, Harley JB, Sestak AL (2005) The mannose-binding lectin gene polymorphisms and systemic lupus erythematosus: two case-control studies and a meta-analysis. Arthritis Rheum 52:3966–3974
Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315:629–634
Higgins JP, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21:1539–1558
Egger M, Smith GD, Phillips AN (1997) Meta-analysis: principles and procedures. BMJ 315:1533–1537
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7:177–188
Huizinga TW, Keijsers V, Yanni G, Hall M, Ramage W, Lanchbury J, Pitzalis C, Drossaers-Bakker WK, Westendorp RG, Breedveld FC, Panayi G, Verweij CL (2000) Are differences in interleukin 10 production associated with joint damage? Rheumatology (Oxford) 39:1180–1188
Tarzi M, Klunker S, Texier C, Verhoef A, Stapel SO, Akdis CA, Maillere B, Kay AB, Larche M (2006) Induction of interleukin-10 and suppressor of cytokine signalling-3 gene expression following peptide immunotherapy. Clin Exp Allergy 36:465–474
Acknowledgments
This study was supported by the Korean Healthcare Technology R&D Project, Ministry for Health, Welfare and Family Affairs, Republic of Korea (grant no. A084794).
Conflict of interest
The authors have no financial or non-financial conflict of interest to declare.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lee, Y.H., Bae, SC., Choi, S.J. et al. Associations between interleukin-10 polymorphisms and susceptibility to rheumatoid arthritis: a meta-analysis. Mol Biol Rep 39, 81–87 (2012). https://doi.org/10.1007/s11033-011-0712-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-011-0712-7