Abstract
Previously published analyses of the association between the interleukin 7 receptor (IL7R) T244I polymorphism (rs6897932) and multiple sclerosis (MS) have yielded conflicting results. We performed a meta-analysis to assess whether the combined data showed this association, and to investigate its effect size. We analyzed 10 studies identified from PubMed (12,185 MS patients and 15,855 controls) and calculated the odds ratios (ORs) and 95% confidence intervals (CIs) for the C-allele, the C/C genotype (recessive effect) and the C/C + C/T (dominant effect) genotype. Heterogeneity within and between studies was observed: allele C: Q = 30.86, P = 0.002; genotype C/C: Q = 30.28, P = 0.003. Using a random-effects model, the C-allele and the C/C genotype were associated with MS (OR = 1.11, 95% CI = 1.04–1.19, P = 0.001 for the C-allele; OR = 1.15, 95% CI = 1.06–1.24, P = 0.0009 for the C/C genotype). The C/C + C/T genotype was also associated with MS using a fixed-effects model (OR = 1.15, 95% CI = 1.05–1.26, P = 0.003). There was no significant publication bias among the selected studies according to the funnel plot. We also performed the analysis on a European subgroup. This revealed an association between IL7R T244I and MS (P < 0.00001 for the C-allele and the C/C genotype; P = 0.0004 for the C/C + C/T genotype), no heterogeneity was observed (allele C: P = 0.07; genotype C/C: P = 0.10). In conclusion, the meta-analysis demonstrated that the IL7R T244I polymorphism was associated with susceptibility to MS.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
MS is an autoimmune-mediated demyelinating disorder characterized by multiple lesions of the central nervous system [1]. The worldwide prevalence of MS is 0.1–0.2%. Generally speaking, the incidence rate among women is more than twice that among men [2]. MS is a complex disease caused by multiple genetic and environmental factors [3, 4]. Although the pathogenesis of MS is not completely understood, a genetic contribution to MS susceptibility has been shown in studies of twins and families, and in genome-wide linkage and association screens [3, 5–9]. Many studies have indicated that the HLA-DRB1 locus on chromosome 6p21 confers susceptibility to MS [10–12]. However, this does not fully explain the genetic basis. Large-scale linkage and association studies have suggested that other loci outside the HLA region have small but non-negligible effects [13].
The association between the IL7R T244I polymorphism and multiple sclerosis is the first non-HLA association widely replicated [14, 15]. The IL7R gene maps to chromosome 5p13 and encodes the interleukin 7 receptor α chain (IL7Rα; also known as CD127) [16]. IL7Rα is a member of the type I cytokine receptor family and forms a receptor complex with the cytokine receptor gamma chain (CD132), for which IL7 is the ligand [17]. It is expressed by cells of the lymphoid lineage, in which it has essential functions in the proliferation and survival of T and B lymphocytes [18]. In MS, the IL7R T244I (rs6897932) single nucleotide polymorphism (SNP) is most likely the causative variant. T244I is located within exon 6 of IL7R, within a transmembrane domain of the encoded protein. IL7R T244I influences the levels of soluble and membrane-bound isoforms of IL7Rα by putatively disrupting an exonic splicing silencer [19]. Some studies have indicated that the ‘C’ allele of IL7R T244I results in an approximately two-fold increase in the skipping of exon 6 compared with transcripts containing the ‘T’ allele, and that this is strongly associated with increased risk of MS [13, 19, 20]. However, others studies have shown a weak or no association between IL7R T244I and susceptibility to MS [21, 22].
We have carried out a meta-analysis of published studies to assess whether the combined evidence demonstrates an association between the IL7R T244I polymorphism and MS, and to perform a preliminary investigation on its effect size.
Materials and methods
Identification of eligible studies and data extraction
Using PubMed, we performed an exhaustive search for all the studies that have examined the association of IL7R T244I with MS. We used the following key words: ‘T244I’, ‘rs6897932’, ‘interleukin 7 receptor’, ‘IL7R’, ‘polymorphism’ and ‘multiple sclerosis’. No restrictions were placed on language, race, ethnicity or geographic area. The following criteria were used to identify relevant published studies: (i) the date of publication was before March 2010; (ii) the data were original (independent studies); (iii) enough information was provided to calculate the odds ratio (OR); (iv) the distribution of genotypes in the control group was in Hardy–Weinberg equilibrium; and (v) the study followed a case–control design and all the controls were healthy individuals. We excluded the following: (i) studies that contained overlapping data; (ii) studies in which the number of wild-type genotypes could not be ascertained; and (iii) studies in which family members had been studied. The following information was extracted from each study: the first author, year of publication, demographics, the number of cases and controls, the P value, the OR or relative risk, the 95% confidence interval (CI) of the OR and the C-allele frequency of the IL7R T244I polymorphism.
Evaluation of the statistical association
We contrasted the effect of the C versus T alleles, the C/C versus the C/T + T/T genotypes (recessive effect) and the C/C + C/T versus the T/T genotypes (dominant effect). The OR and its 95% CI were calculated for each study. In this study, we assessed the within- and between-study variation or heterogeneity by testing Cochran’s Q-statistics [23]. The null hypothesis was that all studies were evaluating the same effect. Not rejecting the above hypothesis usually leads a meta-analysis to adopt a fixed-effects model. The fixed-effects model assumes that the estimated effect sizes only differ by the sampling error. In contrast, if a significant Q-statistic (P < 0.1) indicates heterogeneity across studies, a random-effects model should be adopted [24]. The random-effects model assumes that different studies are measuring different underlying effects and considers both within- and between-study variation.
We measured the degree of inconsistency across studies by calculating the percentage of total between-study variation, because of heterogeneity rather than random variation, as an I 2 metric using the formula: I 2 = Q − d.f./Q, considering I 2 = 1–24% as low heterogeneity; I 2 = 25–49% as moderate heterogeneity, I 2 = 50–74% as large heterogeneity and I 2 > 75% as extreme heterogeneity [25]. Statistical manipulations were performed using the program Review Manager 5.0 (Oxford, UK). We considered the power of each study as the probability of detecting an association between the IL7R T244I polymorphism and MS, and calculated it at the 0.05 level of significance, assuming a small effect size (0.1). The power analysis was performed using G*power (http://www.psycho.uni-duesseldorf.de/aap/projects/gpower).
Results
Studies included in the meta-analysis
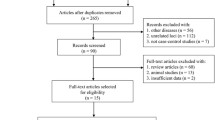
Fourteen association studies related to the IL7R T244I polymorphism and susceptibility to MS were identified through PubMed searches (the most recent article was dated March 2010) [3, 4, 13, 18–22, 26–30]. Four association studies were excluded for overlapping data (the removed studies were [3, 6, 29, 30]). All studies were published in English. Ultimately, ten studies remained for our meta-analysis. A total of 12,185 MS patients and 15,855 controls were investigated (Table 1).
Meta-analysis of available data
In this study we have calculated the combined OR and its 95% CI of the risk C-allele, the C/C and C/C + C/T genotypes. The weighting factors (weight%) used to calculate the combined OR were calculated from the inverse of the variance for each study. Cochran’s Q-statistics and I 2 were used to evaluate heterogeneity between studies. The results of the meta-analysis are shown in Table 2.
Heterogeneity was found among the individual estimates of the ORs for the C-allele and MS (Q = 30.86, P = 0.002, I 2 = 61. Three studies [13, 19, 22] showed that the T-allele was the risk allele, whereas the other studies showed that the C-allele was the risk allele. The risk allele was not consistent across all studies). We therefore adopted the random-effects model to test the association between the IL7R T244I polymorphism C-allele and MS. The overall OR for the C-allele was 1.11, its 95% CI was 1.04–1.19 and the P value was 0.001 (Fig. 1). In other words, the meta-analysis demonstrated that the IL7R T244I C-allele does confer susceptibility to MS.
Heterogeneity was also identified between studies for the C/C genotype and MS (Q = 30.28, P = 0.003, I 2 = 60). We therefore tested the association of the IL7R T244I C/C genotype with MS using a random-effects model. Figure 2 shows that the overall OR was 1.15, its 95% CI was 1.06–1.24 and the P value was 0.0009. This demonstrates that there is an association between the IL7R T244I C/C genotype and susceptibility to MS.
In contrast, there was only a small heterogeneity between studies for the C/C + C/T genotype and MS (Q = 15.73, P = 0.20, I 2 = 24). We therefore adopted the fixed-effects model to test the association between the IL7R T244I polymorphism C/C + T/T genotype and MS. The overall OR was 1.15, its 95% CI was 1.05–1.26 and the P value was 0.003 (Fig. 3). Therefore, the IL7R T244I C/C + C/T genotype was associated with MS in our meta-analysis.
The funnel plot for MS studies showed significant symmetry. It indicated that there was no significant publication bias among the selected studies (Fig. 4).
We have included in the meta-analysis only the studies performed on European population [4, 13, 18, 19, 21, 26, 28]. Under fixed-effects models, the overall OR for the risk C-allele was 1.17 (95% CI = 1.11–1.23, P < 0.00001) (Supplementary Fig. 1), for the recessive effect (C/C genotype) the OR was 1.21 (95% CI = 1.13–1.29, P < 0.00001) (Supplementary Fig. 2), and for the dominant effect (C/C + C/T genotype) the OR was 1.24 (95% CI = 1.10–1.39, P = 0.0004) (Supplementary Fig. 3) (Table 3). In this subgroup, no heterogeneity was observed. However, there was high heterogeneity among all studies. Further studies are required to identify heterogeneity.
Discussion
Since the first association between the IL7R T244I polymorphism and MS was reported in 2003 [14], many studies have attempted to replicate the association. However, these analyses have yielded conflicting results. We performed a meta-analysis to establish the relationship between the IL7R T244I polymorphism and susceptibility to MS. In our meta-analysis, we found a high heterogeneity for the C-allele and the C/C genotype (Q = 30.86, P = 0.002 and Q = 30.28, P = 0.003, respectively) between studies. The common OR for the risk C-allele was 1.11 (95% CI = 1.04–1.19, P = 0.001), for the recessive effect (C/C genotype) it was 1.15 (95% CI = 1.06–1.24, P = 0.0009) and for the dominant effect (C/C + C/T genotype) it was 1.15 (95% CI = 1.05–1.26, P = 0.003). The overall data showed that there is an association between the IL7R T244I polymorphism and MS.
In vitro experiments have shown that T244I affects alternative splicing of exon 6. Transcripts that include exon 6 encode a membrane-bound IL7Rα, whereas transcripts that skip exon 6 produce a predicted soluble form of the protein. Carriers of the ‘C’ allele at T244I have been shown to produce less membrane-bound IL7Rα protein than carriers of the ‘T’ allele, leading to a further increase in the soluble form of IL7Rα. These changes might affect IL7 signaling and enhance the antigenic T cell response to myelin basic protein and myelin oligodendrocyte glycoprotein, both of which have been implicated in the development of MS [19]. Expression of the IL-7–IL7Rα ligand-receptor complex in the cerebrospinal fluid is increased, which supports the simplistic interpretation that increased IL7 signaling induces immune cell proliferation and survival [15]. In summary, the T244I C-allele has been identified as a risk allele for MS in many studies, and this is consistent with our meta-analysis.
The main purpose of performing a meta-analysis is to improve the statistical power and obtain more compelling results by increasing the sample size. However, there are still some limitations. Meta-analyses may be distorted by publication bias and heterogeneity. Our funnel plots showed significant symmetry (Fig. 4), indicating no significant publication bias. Heterogeneity was observed when all studies were included in the meta-analysis, but not within the European subgroup. This suggests that regional differences might be an important reason for the heterogeneity.
References
Mount HT (1973) Multiple sclerosis and other demyelinating diseases. Can Med Assoc J 108(11):1356
Hawkins SA, McDonnell GV (1999) Benign multiple sclerosis? Clinical course, long term follow up, and assessment of prognostic factors. J Neurol Neurosurg Psychiatry 67(2):148–152
Baranzini SE, Wang J, Gibson RA, Galwey N, Naegelin Y, Barkhof F, Radue EW, Lindberg RL, Uitdehaag BM, Johnson MR, Angelakopoulou A, Hall L, Richardson JC, Prinjha RK, Gass A, Geurts JJ, Kragt J, Sombekke M, Vrenken H, Qualley P, Lincoln RR, Gomez R, Caillier SJ, George MF, Mousavi H, Guerrero R, Okuda DT, Cree BA, Green AJ, Waubant E, Goodin DS, Pelletier D, Matthews PM, Hauser SL, Kappos L, Polman CH, Oksenberg JR (2009) Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum Mol Genet 18(4):767–778. doi:10.1093/hmg/ddn388
Weber F, Fontaine B, Cournu-Rebeix I, Kroner A, Knop M, Lutz S, Muller-Sarnowski F, Uhr M, Bettecken T, Kohli M, Ripke S, Ising M, Rieckmann P, Brassat D, Semana G, Babron MC, Mrejen S, Gout C, Lyon-Caen O, Yaouanq J, Edan G, Clanet M, Holsboer F, Clerget-Darpoux F, Muller-Myhsok B (2008) IL2RA and IL7RA genes confer susceptibility for multiple sclerosis in two independent European populations. Genes Immun 9(3):259–263. doi:10.1038/gene.2008.14
Sadovnick AD, Ebers GC, Dyment DA, Risch NJ (1996) Evidence for genetic basis of multiple sclerosis. The Canadian collaborative study group. Lancet 347(9017):1728–1730
Refining genetic associations in multiple sclerosis (2008) Lancet Neurol 7(7):567–569. doi:10.1016/S1474-4422(08)70122-4
Dyment DA, Willer CJ, Scott B, Armstrong H, Ligers A, Hillert J, Paty DW, Hashimoto S, Devonshire V, Hooge J, Kastrukoff L, Oger J, Metz L, Warren S, Hader W, Power C, Auty A, Nath A, Nelson R, Freedman M, Brunet D, Paulseth JE, Rice G, O’Connor P, Duquette P, Lapierre Y, Francis G, Bouchard JP, Murray TJ, Bhan V, Maxner C, Pryse-Phillips W, Stefanelli M, Sadovnick AD, Risch N, Ebers GC (2001) Genetic susceptibility to MS: a second stage analysis in Canadian MS families. Neurogenetics 3(3):145–151
Hermanowski J, Bouzigon E, Forabosco P, Ng MY, Fisher SA, Lewis CM (2007) Meta-analysis of genome-wide linkage studies for multiple sclerosis, using an extended GSMA method. Eur J Hum Genet 15(6):703–710. doi:10.1038/sj.ejhg.5201818
Baranzini SE, Galwey NW, Wang J, Khankhanian P, Lindberg R, Pelletier D, Wu W, Uitdehaag BM, Kappos L, Polman CH, Matthews PM, Hauser SL, Gibson RA, Oksenberg JR, Barnes MR (2009) Pathway and network-based analysis of genome-wide association studies in multiple sclerosis. Hum Mol Genet 18(11):2078–2090. doi:10.1093/hmg/ddp120
Barcellos LF, Sawcer S, Ramsay PP, Baranzini SE, Thomson G, Briggs F, Cree BC, Begovich AB, Villoslada P, Montalban X, Uccelli A, Savettieri G, Lincoln RR, DeLoa C, Haines JL, Pericak-Vance MA, Compston A, Hauser SL, Oksenberg JR (2006) Heterogeneity at the hla-drb1 locus and risk for multiple sclerosis. Hum Mol Genet 15(18):2813–2824. doi:10.1093/hmg/ddl223
Stankovich J, Butzkueven H, Marriott M, Chapman C, Tubridy N, Tait BD, Varney MD, Taylor BV, Foote SJ, Kilpatrick TJ, Rubio JP (2009) Hla-drb1 associations with disease susceptibility and clinical course in Australians with multiple sclerosis. Tissue Antigens 74(1):17–21. doi:10.1111/j.1399-0039.2009.01262.x
Dean G, Yeo TW, Goris A, Taylor CJ, Goodman RS, Elian M, Galea-Debono A, Aquilina A, Felice A, Vella M, Sawcer S, Compston DA (2008) Hla-drb1 and multiple sclerosis in malta. Neurology 70(2):101–105. doi:10.1212/01.wnl.0000284598.98525.d7
Alcina A, Fedetz M, Ndagire D, Fernandez O, Leyva L, Guerrero M, Arnal C, Delgado C, Matesanz F (2008) The T244I variant of the interleukin-7 receptor-alpha gene and multiple sclerosis. Tissue Antigens 72(2):158–161. doi:10.1111/j.1399-0039.2008.01075.x
Teutsch SM, Booth DR, Bennetts BH, Heard RN, Stewart GJ (2003) Identification of 11 novel and common single nucleotide polymorphisms in the interleukin-7 receptor-alpha gene and their associations with multiple sclerosis. Eur J Hum Genet 11(7):509–515. doi:10.1038/sj.ejhg.5200994
Lundmark F, Duvefelt K, Iacobaeus E, Kockum I, Wallstrom E, Khademi M, Oturai A, Ryder LP, Saarela J, Harbo HF, Celius EG, Salter H, Olsson T, Hillert J (2007) Variation in interleukin 7 receptor alpha chain (IL7R) influences risk of multiple sclerosis. Nat Genet 39(9):1108–1113. doi:10.1038/ng2106
Peltonen L (2007) Old suspects found guilty–the first genome profile of multiple sclerosis. N Engl J Med 357(9):927–929. doi:10.1056/NEJMe078147
Noguchi M, Nakamura Y, Russell SM, Ziegler SF, Tsang M, Cao X, Leonard WJ (1993) Interleukin-2 receptor gamma chain: a functional component of the interleukin-7 receptor. Science 262(5141):1877–1880
Akkad DA, Hoffjan S, Petrasch-Parwez E, Beygo J, Gold R, Epplen JT (2009) Variation in the IL7RA and IL2RA genes in german multiple sclerosis patients. J Autoimmun 32(2):110–115. doi:10.1016/j.jaut.2009.01.002
Gregory SG, Schmidt S, Seth P, Oksenberg JR, Hart J, Prokop A, Caillier SJ, Ban M, Goris A, Barcellos LF, Lincoln R, McCauley JL, Sawcer SJ, Compston DA, Dubois B, Hauser SL, Garcia-Blanco MA, Pericak-Vance MA, Haines JL (2007) Interleukin 7 receptor alpha chain (IL7R) shows allelic and functional association with multiple sclerosis. Nat Genet 39(9):1083–1091. doi:10.1038/ng2103
Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, De Jager PL, de Bakker PI, Gabriel SB, Mirel DB, Ivinson AJ, Pericak-Vance MA, Gregory SG, Rioux JD, McCauley JL, Haines JL, Barcellos LF, Cree B, Oksenberg JR, Hauser SL (2007) Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med 357(9):851–862. doi:10.1056/NEJMoa073493
O’Doherty C, Kantarci O, Vandenbroeck K (2008) IL7RA polymorphisms and susceptibility to multiple sclerosis. N Engl J Med 358(7):753–754. doi:10.1056/NEJMc0707553
Rubio JP, Stankovich J, Field J, Tubridy N, Marriott M, Chapman C, Bahlo M, Perera D, Johnson LJ, Tait BD, Varney MD, Speed TP, Taylor BV, Foote SJ, Butzkueven H, Kilpatrick TJ (2008) Replication of KIAA0350, IL2RA, RPL5 and CD58 as multiple sclerosis susceptibility genes in Australians. Genes Immun 9(7):624–630. doi:10.1038/gene.2008.59
Huedo-Medina TB, Sanchez-Meca J, Marin-Martinez F, Botella J (2006) Assessing heterogeneity in meta-analysis: Q statistic or I 2 index? Psychol Methods 11(2):193–206. doi:10.1037/1082-989X.11.2.193
Lau J, Ioannidis JP, Schmid CH (1997) Quantitative synthesis in systematic reviews. Ann Intern Med 127(9):820–826
Zintzaras E (2006) Methylenetetrahydrofolate reductase gene and susceptibility to breast cancer: a meta-analysis. Clin Genet 69(4):327–336. doi:10.1111/j.1399-0004.2006.00605.x
Kallio SP, Jakkula E, Purcell S, Suvela M, Koivisto K, Tienari PJ, Elovaara I, Pirttila T, Reunanen M, Bronnikov D, Viander M, Meri S, Hillert J, Lundmark F, Harbo HF, Lorentzen AR, De Jager PL, Daly MJ, Hafler DA, Palotie A, Peltonen L, Saarela J (2009) Use of a genetic isolate to identify rare disease variants: C7 on 5p associated with MS. Hum Mol Genet 18(9):1670–1683. doi:10.1093/hmg/ddp073
Ramagopalan SV, Anderson C, Sadovnick AD, Ebers GC (2007) Genome wide study of multiple sclerosis. N Engl J Med 357(21):2199–2200. author reply 2200-2191. doi:10.1056/NEJMc072836
Svejgaard A (2008) The immunogenetics of multiple sclerosis. Immunogenetics 60(6):275–286. doi:10.1007/s00251-008-0295-1
Akkad DA (2009) Susceptibility genes for multiple sclerosis. Dissertation, Ruhr-University Bochum
Kallio SP (2009) Novel multiple sclerosis predisposing genetic variants outside the hla region. Dissertation, University of Helsinki
Acknowledgments
This work was supported in part by the National High Tech Development Project of China, the 863 Program (Grant No. 2007AA02Z329), the Natural Science Foundation of Heilongjiang Province (Grant Nos. F2008-02 and D2008-53), the Science Foundation of Heilongjiang Province Education Department (No. 11531113) and the Innovation Fund of Harbin Medical University (No. HCXS2010010).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Ruijie Zhang, Lian Duan, Yongshuai Jiang, Xuehong Zhang—Joint first authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, R., Duan, L., Jiang, Y. et al. Association between the IL7R T244I polymorphism and multiple sclerosis: a meta-analysis. Mol Biol Rep 38, 5079–5084 (2011). https://doi.org/10.1007/s11033-010-0654-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0654-5