Abstract
Ladybird-like genes were recently identified in mammals. The first member characterized, Lbx1, is expressed in developing skeletal muscle and the nervous system. However, little is known about the porcine Lbx1 gene. In the present study, we cloned and characterized Lbx1 from porcine muscle. RT-PCR analyses showed that Lbx1 was highly expressed in porcine skeletal muscle tissues. And we provide the first evidence that Lbx1 has a certain regulated expression pattern during the postnatal period of the porcine skeletal muscle development. Lbx1 gene expressed at higher levels in biceps femoris muscles compared with masseter, semitendinosus and longissimus dorsi muscles in Meishan pigs. Phylogenetic tree was constructed by aligning the amino acid sequences of different species. Moreover, single nucleotide polymorphism (SNP) scanning in the Lbx1 genomic fragment identified two mutations, g.752A>G and g.−1559C>G. Association analysis in our experimental pig populations showed that the mutation of g.752A>G was significantly associated with loin muscle area (P < 0.05) and internal fat rate (P < 0.05). Our results suggest that the Lbx1 gene might be a candidate gene of carcass traits and provide useful information for further studies on its roles in porcine skeletal muscle.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lbx1, a member of the Ladybird-like gene family, encodes a homeodomain transcription factor [1]. Lbx/ladybird genes originated as part of the metazoan cluster of Nk homeobox genes. In all animals investigated so far, both the protostome genes and the vertebrate Lbx1 genes were found to play crucial roles in muscle and neural development [2]. Concerning muscle development, Lbx1 expression is restricted to migrating hypaxial muscle precursor cells and provides thus an excellent marker for the identification of this lineage [3, 4]. Hypaxial muscle forms by two distinct mechanisms: (1) extension of the myotomal sheet for the body wall; (2) migration of individual muscle precursors for limb muscles [3]. The migratory mechanism is also deployed for the formation of the tongue and the diaphragm muscles of mammals [4, 5]. Lbx1 controls the expression of genes that are essential for the recognition or interpretation of cues that guide migrating muscle precursors and maintain their migratory potential [6]. Targets of Lbx1 have not yet been identified in muscle progenitor cells.
The mouse Lbx1 gene is located on chromosome 19 (region D) and the human LBX1 gene maps to the related q24 region of chromosome 10. Jagla et al. [1] showed that mouse Lbx1 and human LBX1 are orthologous genes which have been extremely conserved not only at the level of their protein coding sequence but also in large untranslated regions. Lbx1-deficient mice lack muscles in their limbs due to a defect in migration of muscle precursor cells along a lateral pathway to the limbs [7]. Chick Lbx1 gene is specifically expressed in the prospective hypaxial myoblasts at occipital, cervical and limb levels [3] and overexpression of chick Lbx1 in vivo and in vitro leads to a strong activation of various muscle markers [8]. In addition, the Lbx1 is expressed in a subpopulation of the cardiac neural crest during tubular heart formation. Inactivation of the Lbx1 gene in mice resulted in defects in heart looping, changes in gene expression pattern, and increased cell proliferation ensuing in myocardial hyperplasia [9].
Up until today, the porcine Lbx1 has not been reported. To test the hypothesis that the Lbx1 gene might be the important candidate gene for carcass and growth traits, we cloned the cDNA sequence of porcine Lbx1, analyzed its expression patterns in porcine skeletal muscle. In addition, the genomic sequence, single nucleotide polymorphism (SNP) scanning and association analyses of the porcine Lbx1 gene were also investigated.
Materials and methods
Animals and samples
All pigs in this study were derived from the Experimental Pig Station of Huazhong Agricultural University. They were all fed using standard commercial practices. At eight postnatal periods (3, 21, 35, 60, 90, 120, 150 and 180 days after birth, three samples in each stage), the longissimus dorsi muscles were harvested and then immediately frozen in liquid nitrogen and stored at −80°C. For expression analysis of Lbx1 in different muscles, fast twitch and glycolytic (FG): longissimus dorsi, bicepsfemoris; slow twitch and oxidative (SO): masseter, soleus, semitendinosus were collected from 4-month-old Meishan pigs. Ten different tissues (muscle, ovary, uterus, small intestine, stomach, kidney, lung, spleen, liver, heart) were derived from a 4-month-old Meishan pig and eight different tissues including muscle, intestine, brain, stomach, kidney, lung, liver, heart were collected from a pig fetus (Large White × Meishan F1 hybrid) on 65-day post conception (dpc), for spatial expression analysis. Genomic DNA for mutation polymorphism analysis was prepared from blood using a standard phenol–chloroform method and stored at −20°C.
RNA extraction and cDNA synthesis
Total RNA was isolated from pig tissues with TRIzol Reagent (Invitrogen) according to standard protocols, and the reverse transcription was performed based on the method described previously by Qiao et al. [10].
cDNA and genomic DNA cloning of porcine Lbx1 gene
Based upon the predicted porcine mRNA sequence of Lbx1 gene (GenBank accession No.: XM_001926219), gene-specific primer pairs P1F/P1R and P2F/P2R (Table 1) were designed to amplify porcine Lbx1 cDNA sequence. Two primers (P4F and P4R) (Table 1) were designed to amplify intron of the porcine Lbx1 gene according to the above cDNA sequences we had obtained. In addition, the promoter region of porcine Lbx1 (Genbank accession No.: NC_010456) was amplified by PCR. PCR amplifications were carried out in a 25-μl reaction mixture containing 1 μl (50 ng) of cDNA or DNA as template, 0.5 μl of each primer (5 μM), 2.0 μl of each dNTP (2 mM), 2.5 μl of 10× PCR buffer, 2.0 μl of (25 mmol l−1) Mg2+ and 1 μl (1 U μl−1) of Taq DNA polymerase (Fermentas) and 15.5 μl sterile water. The PCR amplification profiles were as follows: 94°C initial denaturation for 4 min, 35 cycles of 94°C denaturation for 45 s, 57–64°C annealing for 45 s, and 72°C extension for 30–90 s (according the length of the target fragments), followed by a 10 min extension at 72°C. The products were cloned into the pMD 18-T cloning vector (TaKaRa, Dalian, China) and sequenced using M13-forward and M13-reverse primers.
Tissue expression analysis of the porcine Lbx1 gene
The porcine Lbx1 gene mRNA expression in different tissues was detected by RT-PCR with the β-actin gene as an internal control. Lbx1 gene specific primers (LBX1F and LBX1R in Table 1) and β-actin specific primers (β-actinF and β-actinR in Table 1) amplified a product of 143 and 158 bp, respectively. The components for PCR reaction were the same as mentioned above. PCR reactions were conducted under the following condition: initial denaturation at 94°C for 4 min, 30 cycles with 94°C for 40 s, 59°C for 40 s, 72°C for 20 s, followed by a final extension at 72°C for 5 min.
SYBR green real-time RT-PCR analysis of gene expression patterns
The expression level of Lbx1 in eight developmental stages and five different skeletal muscles with different fiber types of Meishan pigs were detected by SYBR Green I assay using ABI 7300 real-time PCR thermal cycle instrument (ABI, USA). Each real-time PCR (in 25 μl) reaction contained 12.5 μl Q-PCR mixture (contains ROX Dye. Toyobo, Jap), 0.25 μM primers (LBX1F and LBX1R in Table 1) and 1 μl normalized template cDNA. The cycling conditions consisted of an initial, single cycle for 3 min at 95°C followed by 40 cycles of cycling consisting of 30 s at 94°C, 30 s at 59°C, 20 s at 72°C, and final extension for 5 min. The specificity of PCR products were confirmed by melting curve analysis. cDNAs from three muscle samples in each stage and fiber types were used to detect the relative expression level of the target gene, and all PCRs were performed in triplicate. Gene expression levels were quantified relatively to the expression of HPRT (HPRT-F and HPRT-R in Table 1) using Gene Expression Macro software (ABI, USA) by employing an optimized comparative Ct(ΔΔCt) value method. Expression levels were considered not detectable when the Ct value of the targeted gene exceeded 35 in the sample tissue. The expression level was calculated as 2(−∆∆Ct) to compare the relative expression. The t test was conducted to identify genes differing in expression, P < 0.05 was considered as significant.
Genome SNP scanning of porcine Lbx1 gene
Five pairs of primers (P3F/P3R, P4F/P4R, P5F/P5R, P6F/P6R and P7F/P7R) were designed to amplify Lbx1 genomic fragment located between −2277 and +1882 with respect to the translation start site (+1). SNPs in the Lbx1 were identified by sequencing PCR products from two pig breeds, including Large White and Meishan.
Allele frequency distribution of the porcine Lbx1 gene in different populations
Animals from four different populations were used to investigate the allele frequency, including 52 Large White pigs, 48 Landrace pigs, 5 Pietrain pigs and 41 Chinese Meishan pigs. Primer pairs P8F/P7R and P9F/P9R were designed to distinguish the SNPs well.
Association analysis of the porcine Lbx1 gene with economic traits
The population used in the association analysis consisted of an F2 population involving Large White and Meishan cross pigs [11]. All F2 pigs were given twice daily diets formulated according to age under a standardized feeding regimen and free access to water. The F2 pigs were slaughtered in 2003 and 2004 following a common protocol [12]. The relationship between genotype and carcass traits was evaluated with the least square method (GLM procedure, SAS version 8.0) using the following statistical model: T ijk = µ + S i + Y j + G k + b ijk X ijk + e ijk , where T ijk is the observed values of traits, µ is the least-square mean, S i is the effect of sex (i = 1 for male or 0 for female), Y j is the effect of year (j = 1 for year 2003 or 0 for year 2004), G k is the effect of genotype (K = AA/CC, AG/CG and GG/GG), b ijk is the regression coefficient of the slaughter weight, X ijk is the slaughter weight, and e ijk is the random residual. According to the method of Liu [13], both additive and dominance effect were also estimated using REG procedure of SAS version 8.0, where the additive effect was denoted as −1, 0 and 1 for AA/CC, AG/CG and GG/GG (TaqI/AccII), respectively, and the dominance effect represented as 1, −1 and 1 for AA/CC, AG/CG and GG/GG (TaqI/AccII), respectively.
Bioinformatic analysis
Open reading frame (ORF) was predicted and the amino acid sequences were deduced with DNAstar software package. Multiple protein sequence alignments were performed using classical ClustalW method (http://www.ebi.ac.uk/clustalw) and the phylogenetic tree was constructed by DNAStar’s LaserGene software (DNAStar, Madison, USA).
Results
PCR amplification and sequence analysis of porcine Lbx1 gene
Analysis of the cDNA and DNA sequence of porcine Lbx1 revealed the following results, (1) this includes two exons and one intron in the porcine genome. The sequence of porcine Lbx1 was deposited in GenBank (GenBank accession No.: HM044216). (2) The cDNA sequence of porcine Lbx1 contains an ORF of 846 bp encoding a protein of 281 residues with a calculated molecular mass of 30.2 kDa and an isoelectric point (PI) of 6.53. A multiple alignment of deduced amino acid sequences of Lbx1 is shown in Fig. 1. The predicted porcine Lbx1 amino acid sequence has 98.5% identity to Bos taurus, 99.2% identity to Canis familiaris, 97.5% identity to Mus musculus, 96% identity to Homo sapiens and 96.4% identity to Pan troglodytes, respectively. Then a phylogenetic tree (Fig. 2) was constructed, which revealed that the porcine Lbx1 has a closer genetic relationship with the Lbx1 of Bos taurus, Canis familiaris, Mus musculus, Homo sapiens, Pan troglodytes than with those of Gallus gallus, Xenopus laevis, Rattus norvegicus. The length of each pair of branches represents the distance between sequence pairs, while the units at the bottom of the tree indicate the number of substitution events.
Multiple amino acid sequence alignments of Lbx1 gene. Protein sequences were obtained from GenBank: XP_001926254 (Ssc: Sus scrofa); XP_614245 (Bta: Bos taurus); XP_543979 (Cfa: Canis familiaris); NP_034821 (Mmu: Mus musculus); NP_006553 (Hsa: Homo sapiens); XP_521589 (Ptr: Pan troglodytes). The symbols (*), (:) and (.) represent completely identical, conservative and semi-conservative amino acid residues, respectively
Tissue expression pattern of porcine Lbx1
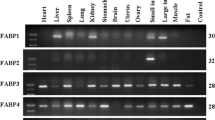
Semi-quantitative RT-PCR was applied to detect the tissue distribution of the porcine Lbx1 gene. The internal control, β-actin, displayed a basically identical signal in each tissue. At 65 dpc, the Lbx1 gene was only expressed in muscle (Fig. 3). At 120 days, the porcine Lbx1 gene was moderately expressed in muscle and heart, weakly in uterus and spleen, and hardly expressed in ovary, small intestine, stomach, kidney, lung and liver (Fig. 4).
Differential expression of Lbx1 gene during skeletal muscle development
SYBR Green real-time PT-PCR analyses revealed that in Meishan pigs, Lbx1 was expressed at 35 days with a relatively high level, and then decreased at 60 days (P < 0.05). No significant expression differences among 60, 90, 120, 150 and 180 days were observed (Fig. 5).
The Lbx1 gene expression in the skeletal muscle from several different stages of Meishan pigs by real-time RT-PCR. Results are averaged from three independent replicates during all stages. Error bars represent SD (n = 3). d3 3 days after birth, d21 21 days after birth, d35 35 days after birth, d60 60 days after birth, d90 90 days after birth, d120 120 days after birth, d150 150 days after birth, d180 180 days after birth. P < 0.05 was considered as significantly different (t test)
Differential expression of Lbx1 gene in five different muscles
To detect the expression pattern of Lbx1 gene in five different muscles, we also performed real-time PCR analyses of samples from three individual 4-month-old Meishan pigs. As shown in Fig. 6, the biceps femoris muscles displayed a greater abundance of Lbx1 mRNA than masseter, semitendinosus and longissimus dorsi muscles in Meishan pigs (P < 0.05). No significant expression differences among masseter, soleus, semitendinosus and longissimus dorsi muscles were observed.
Polymorphism detection
Two mutations, g.752A>G in intron and g.−1559C>G in promoter region of porcine Lbx1 were identified by comparative sequencing. The polymorphism of g.752A>G was analyzed using a TaqI PCR-RFLP, resulting in three fragments (351, 68 and 35 bp) produced by allele G and four fragments (222, 129, 68 and 35 bp) produced by allele A (Fig. 7a). Genetic variation analysis revealed that allele frequencies were significantly different between Chinese Meishan pigs and western commercial pig breeds. The western commercial pig breeds had higher frequencies of the A allele (Table 2).
The polymorphism of g.−1559C>G was analyzed using an AccII PCR-RFLP method. Two different alleles were identified: alleles C (296 bp + 221 bp + 161 bp + 40 bp + 37 bp) and G (457 bp + 221 bp + 40 bp + 37 bp) that control the occurrence of three genotypes, namely: CC, CG and GG (Fig. 7b). The allele distribution indicated that allele G was predominant in the Chinese Meishan pigs. The gene frequencies in different western commercial pig breeds did not show the same trend (Table 3).
Association analysis of the porcine Lbx1 gene with economic traits
The porcine Lbx1 gene g.−1559C>G was not associated with any traits (data not shown). Therefore, another one SNP (g.752A>G) was further used for association analyses with the economic traits. According to the association results, there are significant associations (P < 0.05) between the polymorphism g.752A>G and loin muscle area (LMA), and internal fat rate (IFR) in this F2 population (Table 4).
Discussion
Phylogenetic analyses of Lbx and Lbx-associated genes show that in extant, bony vertebrates only Lbx1- and Lbx2-type genes are maintained. Of these, some Lbx2 sequences evolved faster and were probably subject to neofunctionalisation, while Lbx1 genes may have retained more features of the ancestral Lbx gene [2]. Comparative genomics is the analysis and comparison of genomes from different species. Researchers have learned a great deal about the function of human genes by examining their counterparts in simpler model organisms such as the mouse and some results has revealed that virtually all (99%) of the protein-coding genes in humans align with homologs in mouse, and over 80% are clear 1:1 orthologs [14]. Our results revealed that the porcine Lbx1 gene shares the high sequence identity with its mammalian counterparts at both the nucleotide level and the amino acid level, which suggests the significance and conservation of their biological functions during evolution.
Mouse Lbx1 gene is specifically expressed during embryogenesis, with expression restricted to the developing central nervous system and muscles [1]. In our research, porcine Lbx1 was also found expressed in fetal muscle tissues. In mammalian embryos, myogenic precursor cells emigrate from the ventral lip of the dermomyotome and colonize the limbs, tongue and diaphragm where they differentiate and form skeletal muscle [7]. Lbx1 encodes a homeodomain transcription factor that is expressed exclusively in migrating progenitor cells [15]. This result indicates that porcine Lbx1 gene may play an important role in myogenesis. Subsequently, we examined Lbx1 mRNA spatial expression pattern in a 120-day-old Meishan pig. The Lbx1 mRNA expression distribution in ten tissues displayed that expression in skeletal and heart muscle tissues was higher than other tissues, and relatively lower expression in uterus and spleen. Lbx1 was studied mainly in muscle, however, in the present study it was also found to express in porcine uterus and spleen. The suitable explanation for this was that at the same time those biological activities related to the functions of this gene were presented diversely in different tissues. To explain these expression differences explicitly, further research based on these primary results is needed.
Toward a better understanding of the role of Lbx1 gene in muscle development, we examined the mRNA expression in skeletal muscle of 3, 21, 35, 60, 90, 120, 150 and 180 days of Meishan pigs. The results showed that Lbx1 expression reached its peak about 35 days for Meishan following down-regulation. Postnatal growth is usually considered to be due primarily to muscle cell hypertrophy, but some reports indicate the presence of a third generation of myotubes [16] and a high percentage of proliferative satellite cells after birth [17]. Tertiary myotubes of the pig had been detected in postnatal skeletal muscles [16]. Therefore, hyperplastic mechanisms may still contribute importantly to porcine postnatal growth as recently observed in the rat where new muscle fibres appear in the interstitial space of skeletal muscle [18]. Satellite cells represent the main type of adult stem cells and their proliferation and differentiation increase the postnatal growth of porcine skeletal muscle [17]. Immunohistochemical and northern blot analyses indicated Lbx1 was expressed in activated but not quiescent satellite cells. In vitro, this Lbx1 expression was gradually downregulated when satellite cells differentiate into mature myofibers [19]. Moreover, the percentage of differentiating satellite cells was highest in 1-week-old pigs and significantly decreased in 7-week-old pigs. A slight numerical decline in the percentage of differentiating satellite cells was observed in muscle of pigs between 7 and 21 week of age [17]. It is very interesting that the expression of the porcine Lbx1 gene in skeletal muscle shows the same trend of a higher expression in early postnatal porcine growth. Therefore, we deduce that Lbx1 gene may have a relationship with postnatal hyperplastic mechanisms.
Muscle fiber types and size affect body composition and meat quality traits. According to the amounts of three types of muscle fiber, type I, IIA, and IIB, the properties of skeletal muscle were determined. The three types of muscle fiber differ phenotypically in expressed subsets of myofibrillar isoforms with different adenosine triphosphatase activities as well as different metabolic enzyme activities [20]. Masseter, soleus, and semitendinosus muscles which is composed mostly of type I, slow-twitch oxidative fibres and bicepsfemoris and longissimus dorsi muscle contained predominantly type IIB, fast-twitch glycolytic fibres [21]. When we analyzed the Lbx1 expression in five different muscles, found Lbx1 expressed at higher levels in bicepsfemoris muscle. The differences in the Lbx1 mRNA level between masseter, soleus, semitendinosus and longissimus dorsi muscles were not significant. Thus, our study indicates that there is no direct relationship between the Lbx1 transcription level and type of muscle fiber.
The Lbx1 gene had been assigned to porcine chromosome 14 (SSC14). Several studies in different populations had reported QTLs affecting muscle fiber traits, muscularity and subcutaneous back fat on SSC14 [22, 23]. Therefore, the porcine Lbx1 gene may be an important candidate gene of carcass and growth traits. In this study, the association analysis showed that the loin muscle area and internal fat rate of pigs with AA genotype (g.752A>G) were significantly (P < 0.05) higher than those of pigs with GG genotype (g.752A>G) in this F2 population. The porcine gene g.752A>G does not affect the encoded protein structure. It means that a direct effect of this polymorphism is unlikely. Our results suggest that the associations are caused by linkage disequilibrium with QTLs for these traits. We could not determine whether the g.752A>G is a linkage disequilibrium marker (LD marker) or linkage equilibrium marker (LE marker), so further investigations are required in other pig populations to validate the effect.
In conclusion, our study showed that the Lbx1 gene will be an important candidate gene for carcass traits in animal breeding, and may also be useful for the research of muscle development mechanisms. Our data provide basic molecular information useful for the further investigation of the function of the Lbx1 gene.
References
Jagla K, Dolle P, Mattei MG, Jagla T, Schuhbaur B, Dretzen G, Bellard F, Bellard M (1995) Mouse lbx1 and human lbx1 define a novel mammalian homeobox gene family related to the drosophila lady bird genes. Mech Dev 53(3):345–356
Wotton KR, Weierud FK, Dietrich S, Lewis KE (2008) Comparative genomics of lbx loci reveals conservation of identical lbx ohnologs in bony vertebrates. BMC Evol Biol 8:171. doi 10.1186/1471-2148-8-171
Dietrich S, Schubert FR, Healy C, Sharpe PT, Lumsden A (1998) Specification of the hypaxial musculature. Development 125(12):2235–2249
Dietrich S, Abou-Rebyeh F, Brohmann H, Bladt F, Sonnenberg-Riethmacher E, Yamaai T, Lumsden A, Brand-Saberi B, Birchmeier C (1999) The role of sf/hgf and c-met in the development of skeletal muscle. Development 126(8):1621–1629
Valasek P, Evans DJ, Maina F, Grim M, Patel K (2005) A dual fate of the hindlimb muscle mass: cloacal/perineal musculature develops from leg muscle cells. Development 132(3):447–458. doi:10.1242/dev.01545
Brohmann H, Jagla K, Birchmeier C (2000) The role of lbx1 in migration of muscle precursor cells. Development 127(2):437–445
Gross MK, Moran-Rivard L, Velasquez T, Nakatsu MN, Jagla K, Goulding M (2000) Lbx1 is required for muscle precursor migration along a lateral pathway into the limb. Development 127(2):413–424
Mennerich D, Braun T (2001) Activation of myogenesis by the homeobox gene lbx1 requires cell proliferation. EMBO J 20(24):7174–7183
Schafer K, Neuhaus P, Kruse J, Braun T (2003) The homeobox gene lbx1 specifies a subpopulation of cardiac neural crest necessary for normal heart development. Circ Res 92(1):73–80
Qiao M, Wu HY, Li FE, Jiang SW, Xiong YZ, Deng CY (2009) Molecular characterization, expression profile and association analysis with carcass traits of porcine lcat gene. Mol Biol Reprod 37(5):2227–2234. doi:10.1007/s11033-009-9709-x
Zuo B, Xiong YZ, Deng CY, Su YH, Wang J, Lei MG, Li FE, Jiang SW, Zheng R (2005) Polymorphism, linkage mapping and expression pattern of the porcine skeletal muscle glycogen synthase (gys1) gene. Anim Genet 36(3):254–257. doi:10.1111/j.1365-2052.2005.01286.x
Xiong YZ, Deng CY (1999) Principle and method of swine testing. Chinese Agricultural Press, Beijing
Liu BH (1998) Statistical genomics: linkage, mapping and QTL analysis. CRC Press, LLC, Boca Raton, pp 404–409
An QC, Liu GY (2009) Molecular cloning, sequence identification, and tissue expression profile analysis of three novel porcine genes: Sdhb, snrpa and crybb1. Mol Biol Reprod 36(4):683–690. doi:10.1007/s11033-008-9229-0
Vasyutina E, Stebler J, Brand-Saberi B, Schulz S, Raz E, Birchmeier C (2005) Cxcr4 and gab1 cooperate to control the development of migrating muscle progenitor cells. Genes Dev 19(18):2187–2198. doi:10.1101/gad.346205
Mascarello F, Stecchini ML, Rowlerson A, Ballocchi E (1992) Tertiary myotubes in postnatal growing pig muscle detected by their myosin isoform composition. J Anim Sci 70(6):1806–1813
Mesires NT, Doumit ME (2002) Satellite cell proliferation and differentiation during postnatal growth of porcine skeletal muscle. Am J Physiol Cell Physiol 282(4):C899–C906. doi:10.1152/ajpcell.00341.2001
Tamaki T, Akatsuka A, Ando K, Nakamura Y, Matsuzawa H, Hotta T, Roy RR, Edgerton VR (2002) Identification of myogenic-endothelial progenitor cells in the interstitial spaces of skeletal muscle. J Cell Biol 157(4):571–577. doi:10.1083/jcb.200112106
Watanabe S, Kondo S, Hayasaka M, Hanaoka K (2007) Functional analysis of homeodomain-containing transcription factor lbx1 in satellite cells of mouse skeletal muscle. J Cell Sci 120(23):4178–4187. doi:10.1242/Jcs.011668
Nii M, Hayashi T, Mikawa S, Tani F, Niki A, Mori N, Uchida Y, Fujishima-Kanaya N, Komatsu M, Awata T (2005) Quantitative trait loci mapping for meat quality and muscle fiber traits in a Japanese wild boar × large white intercross. J Anim Sci 83(2):308–315
Karlsson AH, Klont RE, Fernandez X (1999) Skeletal muscle fibres as factors for pork quality. Livest Prod Sci 60(2–3):255–269
Nii M, Hayashi T, Tani F, Niki A, Mori N, Fujishima-Kanaya N, Komatsu M, Aikawa K, Awata T, Mikawa S (2006) Quantitative trait loci mapping for fatty acid composition traits in perirenal and back fat using a Japanese wild boar × large white intercross. Anim Genet 37(4):342–347. doi:10.1111/j.1365-2052.2006.01485.x
Wimmers K, Murani E, Ngu NT, Schellander K, Ponsuksili S (2007) Structural and functional genomics to elucidate the genetic background of microstructural and biophysical muscle properties in the pig. J Anim Breed Genet 124:27–34
Acknowledgments
This study was supported financially by the National “863” project of P. R. China (2007AA10Z166), National High Technology Development Project (2006BAD01A08-01), and Hubei Province Key Project of Science and Technology (2006AA202A01).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chao, Z., Wu, J., Zheng, R. et al. Molecular characterization and expression patterns of Lbx1 in porcine skeletal muscle. Mol Biol Rep 38, 3983–3991 (2011). https://doi.org/10.1007/s11033-010-0516-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0516-1