Abstract
Heat shock protein 90 (HSP90) is not only involved in environmental stress but also plays roles in the ovary development in some vertebrates. To understand its role in crustacean, we examined the HSP90 cDNA for the first time in the ovary and hepatopancreas of the oriental river prawn, Macrobrachium nipponense and designated this protein as MnHSP90 in this study. The MnHSP90 was cloned by the methods of degenerated oligonucleotide primers and rapid amplification of the cDNA ends (RACE). Bioinformatics analysis showed that the MnHSP90 cDNA was 2,684 bp in length, containing a 126 bp 5′ untranslated region (UTR), a 359 bp 3′ UTR, and an open reading frame (ORF) of 2,199 bp encoding a 732-amino acid polypeptide with predicted molecular mass of 84.3 KDa. Sequence alignment showed that the MnHSP90 shared 72–79% identity with other animals. Real-time quantitative PCR (qPCR) analysis demonstrated that the MnHSP90 mRNA was ubiquitously detected in all tested tissues, with the highest expression in the thoracic ganglia, the mediate in heart, muscle and intestine, and the lowest in haemocytes and gills. The MnHSP90 mRNA levels in the hepatopancreas and ovary of M. nipponense reached a maximum at the stage III (early vitellogenic stage) and stage IV (later vitellogenic stage) ovaries, respectively, and then decreased significantly in both tissues as the ovarian development proceeded. The level of MnHSP90 expression in the hepatopancreas was higher than that in the ovary when compared with in the same ovarian developmental stage. Our results indicate that MnHSP90 is involved in ovarian development in oriental river prawn and may play a regulatory role in ovary maturation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Heat shock protein 90 (HSP90) is a major molecular chaperone in cells and has particular significance to the process of cellular regulation. In most cells, 1–2% of cellular proteins are HSP90, making it one of the most abundant proteins in eukaryotes [1, 2]. HSP90 plays crucial roles in protein degradation, protein folding, protein assembly, signal transduction, and myofibril organization in skeletal muscles of embryos under increasing temperature or other stressing conditions [1, 3–10].
Recent studies have shown that HSP90 is also involved in regulating ovarian development in vertebrates. HSP90 binds to estrogen receptor (ER) in the absence of estrogen (E) [11–14] to increase the activity of estrogen hormone-receptor complex to transcribe target genes [15, 16]. In vertebrates, estrogen hormones play a key role in regulating the synthesis of vitellogenin (VTG) [17, 18], the main nutrition for ovary development. During vitellogenesis, estrogen hormones are synthesized in gonad and transported to liver where estrogen (E) and its receptors form a hormone-receptor complex to bind estrogen responsive elements (ERE) located at the upstream of the VTG DNA. This leads to the activation or enhancement of the VTG gene transcription and a subsequent increase and stabilization of VTG mRNA [19]. There is a HSP90-ER-E-VTG regulation channel widely existed in vertebrates [15–19], but it is unclear if the similar mechanism exists in crustacean.
Estrogen hormone has not been found in crustacean for a long time. It is thus believed that there was no HSP90-ER-E-VTG regulation channel in crustacean. However, recent studies have showed that exogenous estrogen hormones can promote ovarian maturation in greasy back shrimp Metapenaeus ensis [20] and kuruma shrimp Marsupenaeus japonicus [21], and accumulation of VTG in M. ensis [22]. Likewise, the level of HSP90 expression in the ovary of crustacean has been reported to be related to the stage of ovarian development [23]. Furthermore, HSP90 was found to be an important regulator participating in VTG synthesis in M. ensis [24]. These findings seem to suggest that the regulatory function of HSP90 in vertebrates may also exist in crustacean. However, little is known on the regulatory function of HSP90 genes in the ovary development of freshwater crustacean. Based the existing information, we hypothesize that HSP90 participates in the regulation of maturation and reproduction in freshwater crustacean.
The oriental river prawn, Macrobrachium nipponense, is a commercially important freshwater prawn in China, Japan, Korea and Vietnam because of its significant contribution to rural economy. The natural populations of M. nipponense in China have declined rapidly due to overfishing, degradation and fragmentation of its natural habitats. Therefore, there is a need to spawn this prawn species under captivity. To achieve a year round seed supply, it is essential to understand the controlling mechanism of ovary maturation. In this study, we aimed to examine the expression of HSP90 in various tissues of this freshwater crustacean and particularly focused on its expression in the ovary and hepatopancreas at different ovarian developmental stages.
Materials and methods
Animals
A total of 170 female oriental river prawns, M. nipponense (1.3–2.1 g) at various ovarian developmental stages were collected from a local seafood market in Shanghai, P. R. China. The developmental stage of ovaries were classified by the criteria described by Gao et al. [25] and Wu et al. [26] (Table 1). A variety of tissues, including heart (HE), gills (GI), muscles (MU), thoracic ganglia (TG), intestine (IN), haemocytes (HA), hepatopancreas and ovary were ablated and stored in liquid nitrogen at −80°C until RNA extraction. The number of animals used for RNA extractions varied among tissues in this study. There were four to six animals used for RNA extractions in ovaries at the ovarian stages I, II and VI, two animals at stage III, and one animal at stages IV and V. One animal was used for the muscle and hepatopancreas, four animals for gills, and 10 animals for the thoracic ganglia, intestine, haemocytes and heart at ovarian stage II. When more than one individual was used, all tissues were pooled for analysis. The husbandry condition throughout the 10-day period was at ambient temperature, under water aeration, and a photoperiod of 12 h light and 12 h dark. The experimental prawns were fed with minced mussels twice daily. Feeding was stopped 12 h before tissue sampling.
Total RNA extraction and reverse transcription
All tissues were homogenized in the Unizol Reagent (Biostar, Shanghai, China), and total RNA was prepared according to the manufacturer’s instruction. Total RNA was quantified on a Genova UV/visible spectrophotometer at 260 nm. The cDNA was synthesized from 5 μg of total RNA by the Takara PrimerScriptTM First Strand cDNA Synthesis kit (TaKaRa, Dalian, China) according to the manufacturer’s instruction.
Characterization of MnHSP90 cDNA
Initially, PCR was performed using the cDNA prepared above as a template, with the degenerated primers of dHSPF and dHSPR (Table 2) based on the highly conserved regions of HSP90 from the alignment of the sequences of Homo sapiens (NM_001017963), Caenorhabditis elegans (NM_074225), Danio rerio (BC065359), Xenopus laevis (AY785160), and Drosophila melanogaster (NM_079175) to obtain the partial fragment of MnHSP90 cDNA.
The cDNA for 5′-RACE and 3′-RACE was synthesized using the Smart Race kit (Clontech, Palo Alto, CA, USA) and the 3′-RACE system (Invitrogen, Carlsbad, CA, USA) respectively, according to the manufacturer’s protocol. A total of 5 μg RNA was reversely transcribed using the MMLV reverse transcriptase (TaKaRa, Dalian, China) with the adapter primer (AP:5′-GGCCACGCG-TCGACTAGTACTTTTTTTTTTTTTTTTT-3′) for 3′-RACE and the 5′-CDS primer A (5′-(T)25 VN-3′, N = A, C, G, or T; V = A, G, or C) for 5′-RACE to obtain the first-strand cDNA. The 5′ and 3′ regions of MnHSP90 were amplified by nested PCR using two 5′-RACE primers (5′ HSPR1, 5′ HSPR2, Table 2) and two 3′-RACE primers (3′ HSPF1, 3′ HSPF2, Table 2) which were based on the known nucleotide sequence of the MnHSP90 cDNA fragment.
The homology search for the nucleotide and protein sequences was performed using BLAST algorithm at NCBI (http://www.ncbi.nlm.nih.gov/). The deduced amino acid sequence was analyzed with the Expert Protein Analysis System (http://www.expasy.org/). The full-length multiple alignment of the MnHSP90 amino acid sequence was compared with the HSP90s of other organisms.
Real time quantitative PCR (qPCR) analysis
The expression of the MnHSP90 mRNA was demonstrated by a SYBR Green real-time quantitative RT-PCR (qPCR) analysis in an ABI StepOne Sequence Detection System. The total RNA for the cDNA synthesis was first digested with RNase-free DNase I to eliminate possible genomic DNA contamination.
The qPCR amplifications were carried out in a total volume of 20 μl, containing 10.0 μl 2× SYBR Premix Ex Taq (TaKaRa, Dalian, China), 2.0 μl diluted cDNA, 0.4 μl 50× ROX reference dye, and 0.5 μl of each primer. The gene-specific primer pairs of HSP90 qHSP90F and qHSP90R (Table 2) were used to amplify the HSP90 transcript. β-actin has been successfully used as an internal gene for the mRNA expression and characterization of the HSP90 gene in M. nipponense [27]. Therefore, the β-actin sequence was used as the internal gene in all qPCR assays, which was amplified with the primers, β-actin F and β-actin R (Table 2) based on the EST sequence (GenBank accession N0: FL589653). The PCR temperature profile was 95°C for 10 s, followed by 40 cycles of 95°C for 10 s, and 60°C for 1 min. DEPC-water for the replacement of template was used as a negative control. Melting curve analysis of amplification products was performed at the end of each PCR reaction to confirm that only one PCR product was amplified and detected.
Data analysis
The MnHSP90 and β-actin standard curves were developed with serially diluted cDNA templates from ovaries of known concentrations (10−1, 10−2, 10−3, 10−4, and 10−5 of the original cDNA solution). The concentration of cDNA in each sample was calculated from the standard curve. The relative expression level of MnHSP90 was calculated by the ratio of the MnHSP90 concentration to the β-actin concentration. The values were imported to Microsoft Excel for subsequent data analyses. All data were presented as means ± SE. The results were subjected to one-way ANOVA test, and the level of significant difference was set at P < 0.05.
Results
Characterization of the MnHSP90 cDNA of M. nipponense
The nucleotide sequence and the deduced amino acid sequence of MnHSP90 are shown in Fig. 1. The sequences included a 126 bp 5′-terminal untranslated region (UTR), a 359 bp 3′ UTR and a 2,199 bp open reading frame (ORF) encoding a 732 amino acid protein polypeptide with predicted molecular mass of 84.3 KDa. In the 3′ UTR, there was one 31 bp poly (A) tail, two polyadenylation signals located 229 bp (ATTTA) and 19 bp (AATAAA), and one upstream poly (A)+ tail.
Nucleotide and deduced amino acid sequences of MnHSP90 from the oriental river prawn. Five amino acid blocks defining the HSP90 protein family and consensus sequence MEED are highlighted as shaded regions. The putative polyadenylation signal site is shown in the open box. The GXXGXG motif is underlined. The RNA instability motif AATAAA is double underlined. The stop codon is indicated by an asterisk
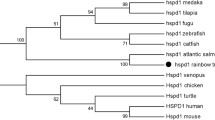
BLAST analysis indicated that MnHSP90 shared significant homology (72–79%) at the nucleotide with the HSP90 sequences reported in Eriocheir sinensis (79%), M. ensis (78%), H. sapiens (74%), Mus musculus (73%) and Opistophthalmus carinatus (72%). The deduced amino acid sequence of MnHSP90 in the oriental river prawn showed a high homology with other crustaceans (E. sinensis 88%, Chiromantes haematocheir 87% and M. ensis 87%), fishes (Salmo salar 77% and Oncorhynchus mykiss 77%), and mammalians (M. musculus 80% and H. sapiens 80%).
Distribution of MnHSP90 mRNA in tissues
The mRNA transcripts of MnHSP90 were widely detected in all examined tissues (Fig. 2a). The highest expression was observed in the thoracic ganglia (TG), which was 2–10 times higher than that in other tissues. MnHSP90 was moderately expressed in muscle (MU), intestine (IN) and heart (HE), while it was least expressed in haemocytes (HA) and gills (GI). There were significant differences of MnHSP90 expression between thoracic ganglia (TG) and other tissues (P < 0.05).
MnHSP90 mRNA expressions in various tissues of the oriental river prawn. a MnHSP90 transcripts in haemocytes (HA), gill (GI), thoracic ganglia (TG), heart (HE), muscle (MU), and intestine (IN) at the ovarian stage II. b MnHSP90 mRNA expression in the hepatopancreas during ovarian development. c MnHSP90 mRNA expression in ovaries during ovarian development. The relative expression of MnHSP90 was measured by the SYBR Green qPCR. Values are shown as mean ± SE (n = 3). Values with the same superscript are not significantly different (P > 0.05)
MnHSP90 cDNA expressions in the ovary and hepatopancreas
The MnHSP90 expressions in the hepatopancreas (Fig. 2b) and ovary (Fig. 2c) were changed as the ovary developed. The relative expressions of MnHSP90 mRNA in the hepatopancreas and ovary reached the maximum at stage III and stage IV, respectively, then dropped with the advance of ovarian development. However, there was no significant difference in MnHSP90 expressions in the ovaries between stage III and IV. And the relative transcript of MnHSP90 mRNA in ovaries was significantly lower than that in the hepatopancreas.
Discussion
The HSP90 acts as a homodimer to facilitate maturation and signal transduction proteins in the regulatory pathway in flatfish Senegalese sole Solea senegalensis Kaup [28]. HSP90 genes have also been isolated from crab [29, 30] and marine shrimp [24, 31], but the expression of HSP90 genes in freshwater crustacean has not been investigated. In the present study, we cloned the full length of the MnHSP90 gene cDNA (GenBank accession N0 GU319963) from a freshwater crustacean species for the first time in an attempt to provide a fundamental basis to understand the molecular mechanism of this protein in regulating the ovary maturation of M. nipponense.
In this study, the deduced amino acid sequence of MnHSP90 included five conserved amino acid motifs which are characterized for the HSP90 protein family signature (i.e., NKEIFLRELISN[S/A]SDALDKIR, LGTIA[K/R]SGT, IGQFGVGFYSA[Y/F]LVA[E/D], IKLYVRRVFI, and GVVDS[E/D]DLPL N[I/V]SRE) [31–33]. In the motif of IGQFGVGFYSA[Y/F]LVA[E/D], the MnHSP90 has Ile14 while the HSP90 in other organisms has Val at the homologous position. This structural similarity is consistent with the HSP90 amino acid of Tigriopus japonicus (ACA03524) and Apis mellifera (XP_395168). We identified a GxxGxG motif in the deduced amino acids, which is essential to the ATP binding in the HSP90 molecular chaperone [34, 35]. The BLAST analysis indicates that the MnHSP90 shares significant homology at the levels of nucleotide (72–79%) and amino acids (78–88%) with HSP90 in other organisms. The conserved characteristics and high similarity with known HSP90s indicate that the MnHSP90 belongs to the HSP90 family. Based on the presence of sequence MEEVD on its C-terminus, the MnHSP90 is concluded to be a cytosolic HSP90 homolog, similar to the HSP90 proteins in other species [36, 37].
MnHSP90 were ubiquitously detected in all examined tissues, with the highest expression in the thoracic ganglia which is a part of the central nervous system in shrimp. The finding can be compared with what found in rabbit [38] and bovine [39] where the highest expressions of HSP90s occur in brain. The current study was the first to report the HSP90 expression in the thoracic ganglia in a crustacean. Interestingly, among the tested tissues, the thoracic ganglia are the only organ reported to secrete hormones to induce ovary maturation in crustacean [40, 41]. In another study, the HSP90 mRNA expression in the ovary of M. ensis starts to increase in stage II, and the level of its expression varies with the stage of ovary development [24]. This result is similar to what we observed on M. nipponense. The results of these two compatible studies suggest that the high expression of HSP90 in the thoracic ganglia supports our hypothesis that HSP90 plays a role in regulating ovary development since the development of thoracic ganglia can stimulate the ovary maturation [40].
Subepidermal adipose tissues, hepatopancreas and ovaries are thought to be the sites for VTG synthesis in crustaceans [42]. The hepatopancreas and ovary are the primary organs for VTG synthesis in oriental river prawn [25]. In this study, we found that the MnHSP90 was detected not only in the ovaries but also in the hepatopancreas. Interestingly, the expression of MnHSP90 mRNA in the hepatopancreas was much higher than that in the ovaries of animals at the same ovarian stage. The hepatopancreas is a gland in crustacean, which combines the digestive functions of the liver and pancreas. However, in the oriental river prawn, the hepatopancreas is proved to be a main organ for VTG synthesis [25], which is consistent with the result in Macrobrachium rosenbergii [43] and Fenneropenaeus chinensis [44].
In M. ensis and Penaeus japonicus, the transcription of VTG in the hepatopancreas and ovaries peaked at a late vitellogenic stage [45–48]. In the present study, the expression of MnHSP90 in these two major organs for VTG synthesis was high in stage III (early vitellogenic stage) and IV (late vitellogenic stage) ovaries but decreased in stage V (mature stage) ovaries. This expression pattern of MnHSP90 in the ovary and hepatopancreas during the ovary development is consistent with that found in M. ensis [24] and P. monodon [23]. Our results showed that the relative transcripts of MnHSP90 mRNA in the ovary and hepatopancreas were related to the stage of ovarian development. The high expression of MnHSP90 prior to the transcription of VTG indicates an active role of HSP90 in the transcriptional regulation of VTG synthesis [24].
The regulatory mechanism of HSP90 for VTG synthesis is controversial. In oviparous vertebrates, there exists a HSP90-ER-E-VTG regulation channel for VTG synthesis. In crustacean, on the other hand, estrogen (E) does not exist. Therefore, the traditional school does not support that a similar regulatory mechanism in oviparous vertebrates exists in crustacean. However, the detection of estrogen in the ovary of crustaceans has challenged the conventional thought in this regard [49, 50]. Wu and Chu [24] reported a strong correlation between estrogen hormones and HSP90 expression in a shrimp M. ensis, suggesting that the expression of VTG may be under the regulation of estrogen hormones through the mechanism similar to that in vertebrates. The detection of MnHSP90 expression in the hepatopancreas and ovary of oriental river prawn supports the hypothesis that HSP90 regulates maturation and reproduction in freshwater crustacean.
References
Picard D (2002) Heat-shock protein 90, a chaperone for folding and regulation. Cell Mol Life Sci 59(10):1640–1648
Csermely P, Schnaider T, Soti C, Prohászka Z, Nardai G (1998) The 90-kDa molecular chaperone family: structure, function, and clinical applications. A comprehensive review. Pharmacol Ther 79(2):129–168
Wiech H, Buchner J, Zimmermann R, Jakob U (1992) Hsp90 chaperones protein folding in vitro. Nature 358(6382):169–170
Miyata Y, Yahara I (1992) The 90-kDa heat shock protein, HSP90, binds and protects casein kinase II from self-aggregation and enhances its kinase activity. J Biol Chem 267(10):7042–7047
Miyata Y, Yahara I (1995) Interaction between casein kinase II and the 90-kDa stress protein, HSP90. Biochemistry 34(25):8123–8129
Schumacher RJ, Hurst R, Sullivan WP, McMahon NJ, Toft DO, Matts RL (1994) ATP-dependent chaperoning activity of reticulocyte lysate. J Biol Chem 269(13):9493–9499
Nathan DF, Vos MH, Lindquist S (1997) In vivo functions of the Saccharomyces cerevisiae Hsp90 chaperone. Proc Natl Acad Sci USA 94(24):12949–12956
Freeman BC, Morimoto RI (1996) The human cytosolic molecular chaperones hsp90, hsp70 (hsc70) and hdj-1 have distinct roles in recognition of a non-native protein and protein refolding. EMBO J 15(12):2969–2979
Du SJ, Li HQ, Bian YH, Zhong YW (2008) Heat-shock protein 90alpha1 is required for organized myofibril assembly in skeletal muscles of zebrafish embryos. Proc Natl Acad Sci USA 105(2):554–559
Yeyati PL, Bancewicz RM, Maule J, Van Heyningen V (2007) Hsp90 selectively modulates phenotype in vertebrate development. PLoS Genet 3(3):0431–0447
Toft DO (1998) Recent advances in the study of hsp90 structure and mechanism of action. Trends Endocrinol Metab 9(6):238–243
Pratt WB, Toft DO (1997) Steroid receptor interactions with heat shock protein and immunophilin chaperones. Endocr Rev 18(3):306–360
Beato M, Klug J (2000) Steroid hormone receptors: an update. Hum Reprod Update 6(3):225–236
Brown MA, Zhu L, Schmidt C, Tucker PW (2007) Hsp90—from signal transduction to cell transformation. Biochem Biophys Res Commun 363(2):241–246
Caruso JA, Laird DW, Batist G (1999) Role of HSP90 in mediating cross-talk between the estrogen receptor and the Ah receptor signal transduction pathways. Biochem Pharmacol 58(9):1395–1403
Fliss AE, Benzeno S, Rao J, Caplan AJ (2000) Control of estrogen receptor ligand binding by hsp90. J Steroid Biochem Mol Biol 72(5):223–230
Soverchia L, Ruggeri B, Palermo F, Mosconi G, Cardinaletti G, Scortichini G, Gatti G, Polzonetti-Magni AM (2005) Modulation of vitellogenin synthesis through estrogen receptor beta-1 in goldfish (Carassius auratus) juveniles exposed to 17-beta estradiol and nonylphenol. Toxicol Appl Pharmacol 209(3):236–243
Orn S, Yamani S, Norrgren L (2006) Comparison of vitellogenin induction, sex ratio, and gonad morphology between zebrafish and Japanese medaka after exposure to 17alpha-ethinylestradiol and 17beta-trenbolone. Arch Environ Contam Toxicol 51(2):237–243
Arukwe A, Goksøyr A (2003) Eggshell and egg yolk proteins in fish: hepatic proteins for the next generation: oogenetic, population, and evolutionary implications of endocrine disruption. Comp Hepatol 2(1):1–21
Yano I (1985) Induced ovarian maturation and spawning in greasy back shrimp, Metapenaeus ensis, by progesterone. Aquaculture 47:223–229
Yano I, Hoshino R (2006) Effects of 17 β-estradiol on the vitellogenin synthesis and oocyte development in the ovary of kuruma prawn (Marsupenaeus japonicus). Comp Biochem Physiol A 144(1):18–23
Tiu SH, Hui JH, He JG, Tobe SS, Chan SM (2006) Characterization of vitellogenin in the shrimp Metapenaeus ensis: expression studies and hormonal regulation of MeVg1 transcription in vitro. Mol Reprod Dev 73(4):424–436
Jiang SG, Qiu LH, Zhou FL, Huang JH, Guo YH, Yang K (2009) Molecular cloning and expression analysis of a heat shock protein (Hsp90) gene from black tiger shrimp (Penaeus monodon). Mol Biol Rep 36(1):127–134
Wu LT, Chu KH (2008) Characterization of heat shock protein 90 in the shrimp Metapenaeus ensis: evidence for its role in the regulation of vitellogenin synthesis. Mol Reprod Dev 75(5):952–959
Gao X, Liu H, Xu J, Cai S (2006) Study on site of vitellogenin synthesis in the freshwater prawn Macrobrachium Nipponese. Biotechnol Bull (z1):437–413 (in Chinese)
Wu P, Qi D, Chen L, Zhang H, Zhang X, Qin J, Hu S (2009) Gene discovery from an ovary cDNA library of oriental river prawn Macrobrachium nipponense by ESTs annotation. Comp Biochem Physiol D 4:111–120
Zhang F, Chen L, Wu P, Zhao W, Li E, Qin J (2009) cDNA cloning and expression of Ubc9 in the developing embryo and ovary of oriental river prawn, Macrobrachium nipponense. Comp Biochem Physiol B 155:288–293
Manchado M, Salas-Leiton E, Infante C, Ponce M, Asensio E, Crespo A, Zuasti E, Cañavate PJ (2008) Molecular characterization, gene expression and transcriptional regulation of cytosolic HSP90 genes in the flatfish Senegalese sole (Solea senegalensis Kaup). Gene 416(1–2):77–84
Li P, Zha J, Zhang ZH, Huang H, Sun HY, Song DX, Zhou KY (2009) Molecular cloning, mRNA expression, and characterization of HSP90 gene from Chinese mitten crab Eriocheir japonica sinensis. Comp Biochem Physiol B 153(3):229–235
Zhang XY, Zhang MZ, Zheng CJ, Liu J, Hu HJ (2009) Identification of two hsp90 genes from the marine crab, Portunus trituberculatus and their specific expression profiles under different environmental conditions. Comp Biochem Physiol C Toxicol Pharmacol 150(4):465–473
Li FH, Luan W, Zhang CS, Zhang JQ, Wang B, Xie YS, Li SH, Xiang JH (2009) Cloning of cytoplasmic heat shock protein 90 (FcHSP90) from Fenneropenaeus chinensis and its expression response to heat shock and hypoxia. Cell Stress Chaperones 14(2):161–172
Buchner J (1999) HSP90 and Co.—a holding for folding. Trends Biochem Sci 24(4):136–141
Caplan AJ (1999) HSP90’s secrets unfold: new insights from structural and functional studies. Trends Cell Biol 9(7):262–268
Prodromou C, Roe SM, O’Brien R, Ladbury JE, Piper PW, Pearl LH (1997) Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell 90(1):65–75
Gupta RS (1995) Phylogenetic analysis of the 90-kD heat shock family of protein sequences and an examination of the relationship among animals, plants, and fungi species. Mol Biol Evol 12(6):1063–1073
Gao Q, Song LS, Ni DJ, Wu LT, Zhang H, Chang YQ (2007) cDNA cloning and mRNA expression of heat shock protein 90 gene in the haemocytes of Zhikong scallop Chlamys farreri. Comp Biochem Physiol B 147(4):704–715
Gao Q, Zhao JM, Song LS, Qiu LM, Yu YD, Zhang H, Ni DJ (2008) Molecular cloning, characterization and expression of heat shock protein 90 gene in the haemocytes of bay scallop Argopecten irradians. Fish Shellfish Immunol 24(4):379–385
Vamvakopoulos NO (1993) Tissue-specific expression of heat shock proteins 70 and 90: potential implication for differential sensitivity of tissues to glucocorticoids. Mol Cell Endocrinol 98(1):49–54
Itoh H, Toyoshima I, Mizunuma H, Kobayashi R, Tashima Y (1990) Three-step purification method and characterization of the bovine brain 90-kDa heat shock protein. Arch Biochem Biophys 282(2):290–296
Jin ZX, Ye HH, Li SJ, Huang HY, Wang GZ (2003) Role of nervous organs in stimulating ovarian maturation in the mud crab Scylla serrata: an in vitro study. Mar Sci 27(1):72–74 (in Chinese)
Kulkarni GK, Glade L, Fingerman M (1991) Oogensis and effects of neuroendocrine tissues on in vitro synthesis of protein by the ovary of the red swamp crayfish, Procambarus clarkii (Girard). J Crustac Biol 11(4):513–522
Meusy JJ, Payen GG (1988) Female reproduction in Malacostracan Crustacea. Zool Sci 5(22):217–265
Yang WJ, Ohira T, Tsutsui N, Subramoniam T, Huong DT, Aida K, Wilder MN (2000) Determination of amino acid sequence and site of mRNA expression of four vitellins in the giant freshwater prawn, Macrobrachium rosenbergii. J Exp Zool 287(6):413–422
Xie S, Sun L, Liu F, Dong B (2009) Molecular characterization and mRNA transcript profile of vitellogenin in Chinese shrimp, Fenneropenaeus chinensis. Mol Biol Rep 36(2):389–397
Tsutsui N, Kawazoe I, Ohira T, Jasmani S, Yang W, Wilder MN, Aida K (2000) Molecular characterization of a cDNA encoding vitellogenin and its expression in the hepatopancreas and ovary during vitellogenesis in the kuruma prawn, Penaeus japonicus. Zool Sci 17(5):651–660
Tsang WS, Quackenbush LS, Chow BK, Tiu SH, He JG, Chan SM (2003) Organization of the shrimp vitellogenin gene: evidence of multiple genes and tissue specific expression by the ovary and hepatopancreas. Gene 303:99–109
Kung SY, Chan SM, Hui JH, Tsang WS, Mak A, He JG (2004) Vitellogenesis in the sand shrimp, Metapenaeus ensis: the contribution from the hepatopancreas-specific vitellogenin gene (MeVg2). Biol Reprod 71(3):863–870
Wong QW, Mak WY, Chu KH (2008) Differential gene expression in hepatopancreas of the shrimp Metapenaeus ensis during ovarian maturation. Mar Biotechnol (NY) 10(1):91–98
Fairs NJ, Quinlan PT, Goad LJ (1990) Changes in ovarian unconjugated and conjugated steroid titers during vitellogenesis in Penaeus monodon. Aquaculture 89(1):83–99
Quinitio ET, Yamauchi K, Hara A, Fuji A (1991) Profiles of progesterone and estradiol-like substances in the hemolymph of female Pandalus kessleri during an annual reproductive cycle. Gen Comp Endocrinol 81(3):343–348
Acknowledgments
This research has been supported by grants from the Chinese Natural Science Foundation (No. 30771670), National Key Project of Scientific and Technical Supporting Programs Funded by Ministry of Science and Technology of China (No. 2006BAD01A13), Zhejiang Key Science and Technology Program of China (No. 2006C12005), Yancheng Institute of Technology (No. XKY2009017) and in part by the E-institute of the Shanghai Municipal Education Commission (Project number E03009). The authors would like to thank Chuanjie Qin for reviewing the manuscript and his helpful suggestions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhao, W., Chen, L., Qin, J. et al. MnHSP90 cDNA characterization and its expression during the ovary development in oriental river prawn, Macrobrachium nipponense . Mol Biol Rep 38, 1399–1406 (2011). https://doi.org/10.1007/s11033-010-0243-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0243-7