Abstract
The NAC family transcription factor has demonstrated its importance in plant development and environmental stress response. Based on the microarray results under salt stress and EST information, the full-length cDNAs of two salt-inducible NAC-family genes (SlNAC1, SlNAM1) were isolated from a salt tolerant tomato cultivar, Edkawi, using Rapid Amplification of cDNA Ends (RACE). SlNAC1 and SlNAM1 encoded 301 and 296 amino acids, respectively, and the deduced protein sequences contained the typical domain of NAC-family transcription factors. Tissue expression profile analysis using semi-quantitative RT-PCR showed that SlNAC1 was expressed mainly in root, flower and green fruit; transcripts of SlNAM1 were detected in all tested tissues except for root, and high-level expression was detected in flower and matured tomato fruit. Both SlNAC1 and SlNAM1 were induced by salt stress in Edkawi, while the expression pattern was different in a salt-sensitive cultivar, ZS-5. Phylogenetic analysis for putative NAC-family peptides available in the tomato genome indicated a wide diversity of this gene family. Results obtained in the present study suggest that both SlNAC1 and SlNAM1 might play important roles in tomato stress tolerance.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
High soil salinity is a serious threat to tomato growth and production. Salinity stress induces the expression of a large number of genes in tomato, including a lot of transcription factor encoding genes [1]. Among the transcription factors, NAC family draws much attention due to its various functions, both in plant development and in abiotic stress tolerance.
The NAC protein family is a widespread while plant-specific transcription factor family, which is identifiable by the presence of a highly conserved N-terminal NAC domain. This domain was originally identified from consensus sequences of petunia NAM, Arabidopsis ATAF1, ATAF2, and CUC2 [2]. Based on decoded genome information, plant genomes usually contain more than 100 NAC-family genes, and they were further classified into diverse subgroups based on sequence similarity (http://plntfdb.bio.uni-potsdam.de). Structure analysis suggests that NAC is a novel type of transcription factor, as the NAC domain does not possess a classical helix-turn-helix motif; instead it consists of a twisted beta-sheet surrounded by a few helical elements [3].
Increasing evidence suggests that NAC family transcription factors play important regulatory roles in various developmental processes and stress responses. NAC proteins are involved in development of the shoot apical meristem, floral organs, lateral shoots, xylary fiber and shoot branching, etc. [2, 4–7]. Direct evidences also reveal that NAC-family genes function in biotic and/or abiotic stress tolerance, including pathogen infection and salt, drought and cold tolerance [8–11]. In the Solanaceae family, the StNAC gene from Solanum tuberosum is induced by Phytophtora infestans infection [12], CaNAC1 gene from Capsicum annuum is up-regulated by bacterial pathogen [13], and SlNAC1 is implicated in the replication of tomato leaf curl virus (TLCV) [14].
In our previous study, two expressed sequence tags (ESTs) annotated as NAC-family genes were found to be salt responsive in a salinity-tolerant tomato cultivar. Here, the molecular cloning of the full-length cDNAs for these two ESTs was presented, and their tissue and salt-inducible expression profile was studied as well.
Materials and methods
Plant materials
Seeds of Edkawi (Tomato Genetic Research Center accession number, LA2711: http://tgrc.ucdavis.edu), a salt-tolerant cultivar, and ZS-5 (Provided by the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences: http://www.ivfcaas.ac.cn), a salt-sensitive commercialized cultivar, were used in this study. Both cultivars belong to Solanum lycopersicum.
For cDNA cloning and gene expression analysis under salt stress, tomato seedlings were grown hydroponically for 35 days (flower bud appearing stage) and then treated with 300 mM NaCl, according to the method of Ouyang et al. [1]. Whole root tissue at 0, 0.25, 0.5, 1, 2, 6, 12, and 24 h after treatment was harvested, immediately frozen in liquid nitrogen, and stored in a deep freezer (−80°C) until total RNA extraction.
For tissue expression profile analysis, plants of Edkawi were grown in soil under normal conditions; tissues of root, stem, young leaf, flower, green fruit and ripe fruit were harvested at the time when the first fruit matured, and stored at −80°C until use.
Methods
Full length cDNA cloning
Based on the previous study, two ESTs (GenBank accession numbers: DY523352, DY523377), annotated as NAC-family genes, were identified as salt stress-inducible genes in Edkawi [1]. To isolate the full length cDNA of the corresponding EST sequences, 5′- and 3′-rapid amplification of cDNA ends (RACE) were performed with cDNA from salt-stressed root. Total RNA samples were isolated from the root tissue of Edkawi under different times of salt stress, as described above using TriZOL (Invitrogen, Carlsbad, CA, USA). Equal amounts of total RNA were pooled and one microgram of the mixed total RNA was reverse transcribed to make 5′- and 3′-RACE-ready first-strand cDNA, according to the instruction of the SMART™ RACE cDNA Amplification Kit (BD Biosciences Clontech, Mountain View, CA, USA). The 5′- and 3′-RACE were carried out in a 25 μl reaction mixtures containing 1× advantage 2 PCR buffer, 0.5 μl 10 mM dNTPs, 2.5 μl BD’s Universal Primer Mix (UPM), 0.5 μl 10 μM gene specific primer, 0.5 μl advantage 2 polymerase (Clontech) and 1 μl 20-fold diluted 5′- and 3′-RACE ready cDNA template. Primers SlNAC1-F and SlNAC1-R were used in 3′- and 5′-RACE for SlNAC1, and primers SlNAM1-F and SlNAM1-R were used in 3′- and 5′-RACE for SlNAM1, respectively (Table S1). The reaction was incubated at 94°C for 1 min followed by 35 cycles of 30 s at 94°C, 30 s at 60°C and 3 min at 72°C, and a final extension at 72°C for 8 min. PCR products were separated in 1% agarose with ethidium bromide (EtBr). Specific amplified DNA bands were recovered using EZ Spin Column DNA Gel Extraction Kit (Bio Basic Inc., Ontario, Canada) and cloned into pMD 18-T according to the manufacturer’s instruction (Takara, Dalian, China). Recombinant clones were sent to Shanghai Sangon Biological Engineering Technology & Services Co., Ltd. for sequencing (Sangon, Shanghai, China). Sequences were assembled into full-length cDNAs based on the 5′ and 3′ fragment information, and the cloned cDNAs were designated SlNAC1 and SlNAM1, respectively.
Genomic DNA sequence obtaining
The genomic DNA sequence of SlNAC1 was retrieved by blasting against tomato genomic sequence (http://solgenomics.net), using cDNA sequence as seed. As to SlNAM1, genomic DNA was extracted from Edkawi seedlings according to the method of Fulton et al. [15] and the genomic DNA of SlNAM1 was amplified using the primers gNAC-F and gNAC-R (Table S1). The PCR product was cloned into pMD 18-T and sequenced as described above. Gene structure diagrams were created using Vector NTI 10 software (Invitrogen) and edited by Adobe Photoshop CS2 (Adobe Systems Inc., Mountain View, CA, USA).
Gene expression analysis under salt stress
Total RNAs were extracted from root tissues at eight time periods (0, 0.25, 0.5, 1, 2, 6, 12, 24 h) for both salt-stressed and control plants of Edkawi and ZS-5 using TriZOL (Invitrogen). Total RNAs were DNase treated and reverse transcription was carried out on five micrograms of total RNA using ReverTra Ace reverse transcriptase (Toyobo, Osaka, Japan) in accordance with the supplier’s manual. RT-PCR analysis was conducted as described [1], with the primers SlNAC1-F and SlNAC1-R for SlNAC1, and the primers SlNAM1-F and SlNAM1-R for SlNAM1 (Table S1). Tomato gene encoding elongation factor 1α was used as internal control (GenBank accession number X53043.1), as a previous report indicates that it appears to be a better internal control under abiotic stress [16]. RT-PCR experiments were repeated three times and the PCR products were visualized in EtBr contained 1% agarose gels. Gel band intensities were quantified by using ImageJ software (National Institutes of Health, Bethesda, MD, USA), and in each gel a relative expression ratio was calculated between the intensity of each band and that of the first band which was amplified from the sample at zero time point.
Tissue expression profile analysis
Total RNAs were isolated from different tissue types of Edkawi as above using TriZOL (Invitrogen). The RT-PCR was the same as described above, except that one microgram of total RNA was used and β-actin gene (GenBank accession number U60478.1) served as the internal control. Band intensities were quantified as above, and the intensity of stem sample for SlNAC1 and intensities of root samples for SlNAM1 and β-actin gene were used for normalization.
Sequence structure and phylogenetic analysis
GENESCAN (MIT, Cambridge, MA) and sequence alignment between cDNA and genomic DNA were used to analyze the exons and introns of genomic DNA. The theoretical molecular weight (Mw) and isoelectric point (pI) were calculated with the ExPASy compute pI/Mw tool [17]. Motif Scan program and Prosite at the ExPASy website (http://www.expacy.ch) were employed to scan for the motifs on the primary structure of the deduced protein sequence [18]. Subcellular location of the putative proteins was predicted with WoLF PSORT [19].
For phylogenetic tree analysis, peptide sequences of putative NAC family transcription factors were retrieved from the Tomato Transcription Factor Database (http://planttfdb.cbi.pku.edu.cn). Together with Senu5 from tomato [20], the deduced proteins described in this study, and 42 characterized NAC family proteins from other plant species, a multiple alignment was carried out using ClustalW [21], and a dendrogram was created using MEGA3 software [22] by the method of Unweighted Pair Group Method with Arithmetic mean (UPGMA) and bootstrap option with 1,000 replications.
Results
Full length cDNA cloning for SlNAC1 and SlNAM1
RACE was applied to clone the full-length cDNAs of two NAC-family genes which were reported as salt-induced genes [1]. The assembled full-length cDNA of one NAC family gene encodes SlNAC1, which was previously reported by Selth et al. [14] The full-length cDNA of SlNAC1 cloned in this study was 1327 bp; it contained a 150 bp of 5′-untranslate region (UTR), which is 22 bp longer than that cloned before [14].
Using cDNA information of SlNAC1, the putative genomic DNA of SlNAC1 was retrieved from the SGN tomato genomic database, the contig containing SlNAC1 is C04.0_contig3, which is located on chromosome 4. The alignment result between the cDNA and genomic DNA showed that SlNAC1 contained three exons and two introns. The gene structure of SlNAC1 was shown in Fig. S1a.
Sequence analysis showed that the full-length cDNA for SlNAM1 was 1218 bp, including a 103 bp 5′-UTR and 224 bp 3′-UTR, respectively. The intact open reading frame encoded 296 amino acids. Genomic DNA of SlNAM1 was cloned from Edkawi and sequence alignment between the genomic DNA and cDNA showed three exons and two introns were existed in this gene (Fig. S1b).
The related sequences were all submitted to GenBank (www.ncbi.nlm.nih.gov). The accession numbers are: EU670749.1 (SlNAM1 full-length cDNA), EU670750.1 (SlNAC1 full-length cDNA), and GU256056 (SlNAM1 genomic DNA).
Putative protein feature of SlNAM1 and SlNAC1
The estimated molecular weight was 34.8 kDa for SlNAC1 and 33.9 kDa for SlNAM1 and their pI were predicted as 7.62 and 6.76, respectively. MotifScan results showed that a conserved NAC domain (aa13-163) and a NAM domain (aa13-138) located in SlNAC1, and NAC (aa8-159) and NAM (aa8-133) domain were also detected in SlNAM1. According to the subcellular location prediction results, both proteins were located in the nucleus.
Tissue expression profiles for SlNAM1 and SlNAC1
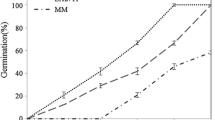
Semi-quantitative RT-PCR results showed that the expression level of SlNAM1 was very high in tomato flower tissue and ripe fruit, while it was very low in root tissue. The transcript for SlNAC1 was abundant in root tissue, whereas its level was relatively lower in other tissues, especially in stem and matured fruit (Fig. 1, Table S2).
Expression of SlNAM1 and SlNAC1 in Edkawi and ZS-5 under salt stress
Different expression patterns upon salt stress for SlNAM1 and SlNAC1 were revealed in salt-tolerant and salt-sensitive tomato genotypes, using semi-quantitative RT-PCR analysis (Fig. 2, Table S3). SlNAM1 was induced in both the salt-tolerant cultivar, Edkawi, and the salt-sensitive cultivar, ZS-5, while the induction was sooner in Edkawi. Transcripts increased within 30 min under salt stress in Edkawi; however, the increase was detected after 6 h in ZS-5. The expression level of SlNAM1 was very stable in the non-stressed control of both genotypes.
Expression of SlNAM1 and SlNAC1 in Edkawi and ZS-5 upon 300 mM salt stress. The number on the top indicates different time points (h) after treatment. Number of PCR cycles (C) is listed on the right side. Plants were grown hydroponically for 35 days and then treated with 300 mM NaCl solution, and samples from whole root tissue were taken for analysis. Tomato gene encoding elongation factor 1α was used as internal control
Compared to SlNAM1, SlNAC1 showed a slow induction pattern upon salt stress in Edkawi, while its expression in ZS-5 was distinct. The expression of SlNAC1 in ZS-5 was stronger in 15 min, 1 h, and 6 h. Interestingly, in both Edkawi and ZS-5 control plants, the expression of SlNAC1 was high during 0–1 h after salt treatment and then it was down-regulated in the later time points.
Discussion
In this study, we cloned two NAC-family genes and investigated their tissue expression profiles and temporal patterns of induction upon salt stress.
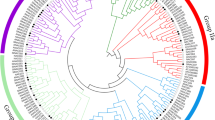
The tissue and salt-stress expression pattern of the two genes were different, reflecting functional diversification of tomato NAC-family genes. To further explore this phenomenon, all the putative NAC family transcription factors were retrieved from the Tomato Transcription Factor Database. Only 41 putative NAC family peptides were obtained from the database, and alignment results showed that PTLe00691.1 and PTLe00692.1 were duplicate records. All these putative transcription factors, together with the previously identified Senu5 [20], SlNAC1 and SlNAM1 described here and 42 characterized NAC family members from other plant species were aligned and the phylogenetic tree was constructed (Fig. 3). The tomato NAC-family transcription factors were classified into more than 12 groups: ANAC042, ATAF1, NAM, NAM-B1, NAP, NTM, Senu5, StNAC, Stress NAC, TERN, TIP and VND. And SlNAM1 belonged to the type of ATAF1, while SlNAC1 belonged to the StNAC type. This phylogenetic tree analysis indicated the extremely functional diversification of NAC gene family in tomato. With the progress of the international tomato genome sequencing project (www.sgn.cornell.edu), more NAC-family genes are to be identified. The number of NAC would be expected to exceed those of Arabidopsis or rice. A detailed expression analysis of these genes under different environmental conditions (i.e., biotic stress, abiotic stress, nutrition deficiency) would help us to speculate on their biological functions.
Phylogenetic tree of NAC-family peptides from tomato. The multiple alignment was made with ClustalW and the dendrogram was built with the MEGA3 software, as a consensus of 1,000 bootstrap replicates by the UPGMA method (numbers at nodes indicate the percentage bootstrap scores). The subfamilies within the NAC superfamily, as designated by Ooka et al. [27], are grouped by different colors. Arrows indicate the two NAC-family genes described in this study. Besides the Senu5 (CAA99760) identified in tomato, NAC proteins from other plant species were also included in this analysis, whose accession numbers are: ANAC019 (NP_175697), ANAC028 (NP_176766.1), ANAC042 (NP_181828.1), ANAC055 (NP_188169), ANAC072 (NP_001078452), ATAF1 (NP_171677), ATAF2 (NP_680161), AtNAC1 (AAF21437), AtNac2 (AAO41710), AtNAC3 (AAP42729), BnNac1-1 (AAP35048), BnNac5-1 (AAP35050), BnNac5-7 (AAP35051), BnNac5-8 (AAP35052), BnNac5-11 (AAP35053), BnNac14 (AY245886), BnNac18 (AAP35054), BnNac485 (AAP35056), CaNAC1 (AAW48094), CUC1 (BAB20598), CUC2 (BAA19529), CUC3 (AAP82630), GRAB1 (CAA09371), GRAB2 (CAA09372), NAM (CAA63102), NAM-B1 (CAG28971), NAP (AF402603_1), NST1 (ANAC043), NTL9 (NP_001119122.1), Os02g0745300 (NP_001048109.1), OsNAC3 (BAA89797), OsNAC4 (AB028183), OsNAC6 (BAA89800), OsNAC19 (AY596808), PetNAM3 (AF509866.1), PetNAM10 (AF509873), StNAC (CAC42087), TERN (BAA78417.1), TIP (AAM47025.1), VND1 (NP_179397.1), VND2 (NP_195339.1) and VND4 (NP_172690.1). Abbreviations for the name of the subfamilies are: ANAC042 Arabidopsis NAC protein 042 transcription factor like family, ATAF1: Arabidopsis transcription factor 1 like family, ATAF2: Arabidopsis transcription factor 2 like family, NAM no apical meristem transcription factor like family, NAM-B1 no apical meristem B1 transcription factor like family, NAP NAC-like activated by APETALA3/PISTILLATA family, NTM NAC with transmembrane motif transcription factor family, Senu5 Tomato senescence up-regulated 5 like family, StNAC: Solanum tuberosum NAC transcription factor like family, Stress NAC stress NAC transcription factor family, TERN Tobacco elicitor-responsive NAC protein like family, TIP turnip crinkle virus interacting protein like family, VND vascular-related NAC domain family
SlNAC1 was expected to function in both biotic and abiotic stress. SlNAC1 was reported to be involved in the interaction with an geminivirus replication enhancer (REn) protein and plays an important role in the process of REn enhanced TLCV replication [14]. SlNAC1 is induced by TLCV, while this gene was also induced by salt stress according to our results. SlNAC1 has high homology with CaNAC1 and StNAC. CaNAC1 from pepper was activated by pathogen, salicylic acid and ethylene, indicating its role in defense [13]. StNAC from potato is induced by pathogen infection and wounding [12]. Also, SlNAC1 and rice OsNAC6 belonged to the same subgroup in the phylogenetic analysis. OsNAC6 is induced by cold, salt, drought and abscisic acid etc. [23]. This suggested that SlNAC1 might play an important role in the cross talk of different kinds of stresses.
The expression pattern of SlNAC1 was interesting. SlNAC1 was induced in Edkawi under salt stress, but it was declined without salt treatment. In the salt-sensitive tomato cultivar ZS-5, SlNAC1 showed a wavy pattern, with higher expression at 15 min, 1 h, and 6 h time points. While under control conditions, its expression pattern is similar to that of Edkawi. With the availability of the genomic data from tomato cultivar Heinz 1706 (Solanum lycopersicum), the 1.5 kb promoter region of SlNAC1 was retrieved from the genomic sequence. Cis-acting regulatory element analysis in the promoter sequence using PlantCARE [24] showed that there are two abscisic acid response elements, one anaerobic response element, various light responsive elements, two heat shock elements, one low-temperature-response element, two MYB binding sites and four circadian control elements in the promoter. Other elements such as MeJA-responsiveness, salicylic acid responsiveness, fungal elicitor responsive element were also found in the promoter. The surprising pattern of SlNAC1 in ZS-5 might be due to the complex interaction between salt stress and other factors such as anaerobic (although continuous aeration was provided before and after the salt stress treatment), heat, temperature change, and circadian. And salt response in salt-tolerant cultivar Edkawi seems to be the dominant effect in this scenario.
It was expected that SlNAM1 might have multiple functions in both development and stress response. Homology analysis showed that SlNAM1 has relative higher identity with Arabidopsis ATAF1 and Brassica napus BnNac14. Lines of evidence have shown that these genes function in development and biotic/abiotic stress response [25, 26]. The RT-PCR results in this study also supported this speculation. SlNAM1 was highly expressed in flower tissue and mature fruit, indicating its potential role in organ development. And the salt-inducible expression of this gene suggested its possible role in stress tolerance. Currently, the functional characterization of the cloned NAC-family genes is undergoing study.
References
Ouyang B, Yang T, Li H, Zhang L, Zhang Y, Zhang J, Fei Z, Ye Z (2007) Identification of early salt stress response genes in tomato root by suppression subtractive hybridization and microarray analysis. J Exp Bot 58:507–520. doi:10.1093/jxb/erl258
Aida M, Ishida T, Fukaki H, Fujisawa H, Tasaka M (1997) Genes involved in organ separation in Arabidopsis: an analysis of the cup-shaped cotyledon mutant. Plant Cell 9:841–857. doi:10.1105/tpc.9.6.841
Ernst HA, Olsen AN, Larsen S (2004) Structure of the conserved domain of ANAC, a member of the NAC family of transcription factors. EMBO Rep 5:297–303. doi:10.1038/sj.embor.7400093
Sablowski RW, Meyerowitz EM (1998) A homolog of NO APICAL MERISTEM is an immediate target of the floral homeotic genes APETALA3/PISTILLATA. Cell 92:93–103. doi:10.1016/S0092-8674(00)80902-2
Xie Q, Frugis G, Colgan D, Chua NH (2000) Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes Dev 14:3024–3036. doi:10.1101/gad.852200
Ko JH, Yang SH, Park AH, Lerouxel O, Han KH (2007) ANAC012, a member of the plant-specific NAC transcription factor family, negatively regulates xylary fiber development in Arabidopsis thaliana. Plant J 50:1035–1048. doi:10.1111/j.1365-313X.2007.03109.x
Mao C, Ding W, Wu Y, Yu J, He X, Shou H, Wu P (2007) Overexpression of a NAC-domain protein promotes shoot branching in rice. New Phytol 176:288–298. doi:10.1111/j.1469-8137.2007.02177.x
Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L (2006) Overexpressing a NAM, ATAF and CUC (NAC) transcription factor enhances drought resistant and salt tolerance in rice. Proc Natl Acad Sci USA 103:12987–12992. doi:10.1073/pnas.0604882103
Bu Q, Jiang H, Li CB, Zhai Q, Zhang J, Wu X, Sun J, Xie Q, Li C (2008) Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Res 18:756–767. doi:10.1038/cr.2008.53
Hu H, You J, Fang Y, Zhu X, Qi Z, Xiong L (2008) Characterization of transcription factor gene SNAC2 conferring cold and salt tolerance in rice. Plant Mol Biol 67:169–181. doi:10.1007/s11103-008-9309-5
Yokotani N, Ichikawa T, Kondou Y, Matsui M, Hirochika H, Iwabuchi M, Oda K (2009) Tolerance to various environmental stresses conferred by the salt-responsive rice gene ONAC063 in transgenic Arabidopsis. Planta 229:1065–1075. doi:10.1007/s00425-009-0895-5
Collinge M, Boller T (2001) Differential induction of two potato genes, Stprx2 and StNAC, in response to infection by Phytophthora infestans and to wounding. Plant Mol Biol 46:521–529. doi:10.1023/A:1010639225091
Oh SK, Lee S, Yu SH, Choi D (2005) Expression of a novel NAC domain-containing transcription factor (CaNAC1) is preferentially associated with incompatible interactions between chili pepper and pathogens. Planta 222:876–887. doi:10.1007/s00425-005-0030-1
Selth LA, Dogra SC, Rasheed MS, Healy H, Randles JW, Rezaian MA (2005) A NAC domain protein interacts with tomato leaf curl virus replication accessory protein and enhances viral replication. Plant Cell 17:311–325. doi:10.1105/tpc.104.027235
Fulton TM, Chunwongse J, Tanksley SD (1995) Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol Biol Rep 13:207–209. doi:10.1007/BF02670897
Nicot N, Hausman JF, Hoffmann L, Evers D (2005) Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot 56:2907–2914. doi:10.1093/jxb/eri285
Bjellqvist B, Hughes GJ, Pasquali C, Paquet N, Ravier F, Sanchez JC, Frutiger S, Hochstrasser D (1993) The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 14:1023–1031. doi:10.1002/elps.11501401163
Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJ (2008) The 20 years of PROSITE. Nucleic Acids Res 36:D245–D249. doi:10.1093/nar/gkm977
Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res 35:W585–W587. doi:10.1093/nar/gkm259
John I, Hackett R, Cooper W, Drake R, Farrell A, Grierson D (1997) Cloning and characterization of tomato leaf senescence-related cDNAs. Plant Mol Biol 33:641–651. doi:10.1023/A:1005746831643
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. doi:10.1007/978-1-4020-6754-9_3188
Kumar S, Nei M, Dudley J, Tamura K (2008) MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinform 9:299–306. doi:10.1093/bib/bbn017
Ohnishi T, Sugahara S, Yamada T (2005) OsNAC6, a member of the NAC Gene family, is induced by various stresses in rice. Genes Genet Syst 80:135–139. doi:10.1266/ggs.80.135
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327. doi:10.1093/nar/30.1.325
Hegedus D, Yu M, Baldwin D, Gruber M, Sharpe A, Parkin I, Whitwill S, Lydiate D (2003) Molecular characterization of Brassica napus NAC domain transcriptional activators induced in response to biotic and abiotic stress. Plant Mol Biol 53:383–397. doi:10.1023/B:PLAN.0000006944.61384.11
Wu Y, Deng Z, Lai J, Zhang Y, Yang C, Yin B, Zhao Q, Zhang L, Li Y, Yang C, Xie Q (2009) Dual function of Arabidopsis ATAF1 in abiotic and biotic stress responses. Cell Res 19:1279–1290. doi:10.1038/cr.2009.108
Ooka H, Satoh K, Doi K, Nagata T, Otomo Y, Murakami K, Matsubara K, Osato N, Kawai J, Carninci P, Hayashizaki Y, Suzuki K, Kojima K, Takahara Y, Yamamoto K, Kikuchi S (2003) Comprehensive analysis of NAC family genes in Oryza sativa and Arabidopsis thaliana. DNA Res 10:239–247. doi:10.1093/dnares/10.6.239
Acknowledgments
This work was supported by the Natural Science Foundation of China (No. 30400299 and No. 30771461).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11033_2010_177_MOESM1_ESM.jpg
Gene structure of SlNAC1 (a) and SlNAM1 (b). Exons are represented as black boxes. Numbers in boxes indicate exon size in base pairs (bp). Introns are shown as lines linking exons and their sizes are shown in base pairs. The start codon is shown as ATG, and stop codon is TAA for SlNAC1 and TGA for SlNAM1. NAC or NAM domain is shown as line or dot line below the exons, and numbers on the left indicate amino acid positions of the protein domains (JPG 550 kb)
Rights and permissions
About this article
Cite this article
Yang, R., Deng, C., Ouyang, B. et al. Molecular analysis of two salt-responsive NAC-family genes and their expression analysis in tomato. Mol Biol Rep 38, 857–863 (2011). https://doi.org/10.1007/s11033-010-0177-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0177-0