Abstract
Finger millet (Eleusine coracana L.) is an important crop used for food, forage, and industrial products. Three DNA marker techniques, random amplified polymorphic DNA (RAPD), simple sequence repeat (SSR) and cytochrome P450 gene based markers were used for the detection of genetic polymorphism in 83 accessions of finger millet collected from various geographical regions of India and Africa. A total of 18 RAPD, 10 SSR and 10 pairs of cytochrome P450 gene based markers were generated 56.17, 70.19 and 54.29% polymorphism, respectively. Mean polymorphism information content (PIC) for each of these marker systems (0.280 for RAPD, 0.89 for SSR and 0.327 for cytochrome P450 gene based markers) suggested that SSR marker were highly effective in determining polymorphism. The phenograms based on the three markers data indicate that genotypes from different geographical regions are clearly distinguishable as separate clusters. Mantel test employed for detection of goodness of fit established cophenetic correlation values above 0.90 for all the three marker systems. The dendrograms and PCA plots derived from the binary data matrices of the three marker systems are highly concordant. High bootstrap values were obtained at major nodes of phenograms through WINBOOT software. Based on the results of present study, SSR and cytochrome P450 gene based markers appear to be particularly useful for the estimation of genetic diversity. This study reveals the potential of RAPD, SSR and gene based markers for characterizing germplasm of Eleusine coracana and narrow down the vast germplasm into distinct core groups.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Eleusine coracana, commonly called finger millet and ragi, is an allotetraploid cereal, is widely cultivated in the arid and semiarid regions of the world. It is rich in protein (~6–13%) and calcium (~0.3–0.4%) [1] and serves as an important staple food for rural populations in developing tropical countries where calcium deficiency and anaemia are widespread [2]. On the other hand, some of the health benefits such as hypoglycemic, hypocholestrolemic and anti-ulcerative are associated with the millet [3]. DNA fingerprinting techniques are very useful in correct identification of taxa. The most commonly used marker systems are random amplified polymorphic DNA (RAPD) [4, 5], restriction fragment length polymorphism (RFLP) [6], amplified fragment length polymorphism (AFLP) [7], inter simple sequence repeats (ISSRs) [8] and microsatellites or simple sequence repeats (SSRs) [9]. Randomly amplified polymorphic DNA (RAPDs) have been extensively used for the assessment of genetic diversity in a variety of plants like Zea mays [10], barley [11], rice [12, 13], wheat [14], etc. Random Amplified Polymorphic DNA analysis is quick [15], however, problems with reproducibility in amplification of RAPD markers and with data scoring have been reported. SSRs are multi-allelic; they have high potential for use in evolutionary studies and studies regarding genetic diversity and relationships analysis in a variety of plants like wheat [16], sorghum [17] and Zea mays [18], etc. Cytochrome P450 mono-oxygenases are widely found in animals, plants and microorganisms [19]. Sequence diversity of P450 gene-analogues in different plant species have been studied and it has been reported that P450 gene-analogues can be used as new genetic markers for diversity studies in plants reflecting both functional and genome-wide regions [20, 21]. Data mining on Arabidopsis thaliana has resulted in the development of number of primer-sets derived from Cyt P450 genes, which could be used as universal tools for the assessment of genome-wide diversity in diverse plant species lacking relevant genetic markers [22].
Assessment of genetic diversity in finger millet (Eleusine coracana L.) was done using random amplified polymorphic DNA (RAPD) [23–25], restriction fragment length polymorphism (RFLP) [26], and interspersed simple sequence repeat (ISSR) markers [23, 27], isozymes [27] and SSRs [28, 29]. Recently attempts have been made to develop genetic map of tetraploid finger millet (Eleusine coracana subsp. coracana) genome (2n = 4x = 36) using RFLP, AFLP, EST and SSR markers [29]. The availability of genetic map of E. coracana could be used to investigate the diversity of different genes and its possible locations to specific chromosomes in near future.
We have conducted this study to analyze genetic diversity present in the accessions of Eleusine coracana collected from different agro-climatic regions of India and Africa. We have used RAPD, SSR and Cytochrome P450 gene based markers for this purpose and the potential of these molecular markers was also compared.
Materials and methods
Plant material
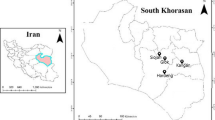
A total of 83 accessions were used in the present study. Seed samples of 83 accessions of Eleusine coracana (collected from different regions of India and Africa (Map 1) were obtained from, Project Coordination Cell, All India Co-ordinated Small Millets Improvement Project, ICAR, UAS, GKVK, Bangalore. The pass port data was available with the accessions (Table S1).
DNA extraction
The genomic DNA of different accessions of finger millets was isolated by standard method [30] quantified and analyzed on agarose gel electrophoresis [31].
RAPD, SSR and Cytochrome P450 gene based markers analyses
A total of 18 random primers, 10 SSR primers and 10 pairs of cytochrome P450 gene based primers were used for the polymorphism survey. Randomly amplified polymorphic DNA assay was carried out in 25 μl reaction volume containing 50 ng DNA, 5 mM MgCl2, 2 mM each of dNTP, 10 μM random primers, 0.6 U Taq polymerase (Bangalore Genei pvt. Bangalore, India). Amplification was performed with thermal cycler (Eppendorf Germany).The standardized amplification was: Initial denaturation 95°C for 5 min. followed by 40 cycles of denaturation 94°C for 1 min; Primer annealing based on T m value for 1 min; primer extension at 72°C for 2 min; and final primer extension at 72°C for 7 min. For SSR analysis, 50 ng of template DNA was subjected to amplification with microsatellite primers, other components remaining same as that of RAPD. The annealing temperatures of the cycling parameter were readjusted for each microsatellite primers according to their calculated melting temperature (T m) based on the sequence composition [T m = 4° (G+C) + 2° (A+T) − 3°C]. For Cytochrome P450 gene based markers PCR amplification was performed as per the standard protocol using 50 ng of template DNA, 5 mM MgCl2, 2 mM each of dNTP, 10 μM of forward and reverse primers, 1.5 U Taq polymerase (Bangalore Genei pvt. Bangalore, India) in a volume of 25 μl.
PCR amplified products of all the primers were subjected to gel electrophoresis using 1.8% agarose gel in 1× TBE buffer. The fragment sizes, ranged from 0.3 to 4.0 kb were detected by comparing the amplicons with a 100 bp DNA ladder and EcoR1/HindIII double digest λ-DNA as marker (Genei Pvt., Bangalore, India) and the ethidium bromide stained gels were documented using Alpha Imager 1200TM (Alpha Innotech Corporation, USA). Duplicated independent DNA preparations for each sample were done and only major bands consistently amplified were scored.
Data analysis
The amplified products were scored for the presence (1) or absence (0) of bands of various sizes across the genotypes to generate a binary matrix. Data were analyzed using the NTSYS-pc version 2.11w software to calculate the similarity values and generate the phenogram [32]. The SIMQUAL program was used to calculate the Jaccard’s coefficients. Jaccard’s similarity coefficient was utilized for estimating the pairwise similarity between the operational taxonomic units (OTUs) using the formula Jaccard’s coefficient = N AB/(N AB + N A + N B) where, N AB is the number of bands shared by samples, N A represents amplified fragments in sample A, and N B represents fragments in sample B [33]. Similarity matrices were utilized to construct the UPGMA (unweighted pair- group method with arithmetic average) dendrograms, clustering was performed by sequential agglomerative hiearchical nested clustering, a distance based method, where series of successive mergers are used to group individuals with similar characteristics.
The product-moment correlation (r) based on Mantel Z-value [34] was computed to measure the degree of relationship between similarity index matrices produced by any two-marker systems. To determine robustness of the dendrogram, the data were bootstrapped with 1,000 replications along with Jaccard’s coefficient by the computer programme WINBOOT [35].
Polymorphic information content (PIC) was calculated using the formula PIC = 1 − ∑p 2 i , where p i is the frequency of the ith allele. Average heterozygosity (Hav) is obtained by taking the average of PIC values obtained for all the markers. Multiplex ratio (MR) for each assay was estimated by dividing the total number of bands (monomorphic—m, and polymorphic—p) amplified by the total number of assays (primer combinations employed—n) as per [36].
Marker index (MI) was obtained by multiplying the average heterozygosity (Hav) with MR.
Results
RAPD analysis
Randomly amplified polymorphic DNA assays were performed with a total of 18 random primers. Initial screening of 18 RAPD primers on 83 accessions showed that 7 of these were monomorphic. In total, 90 amplicons were scored in the size range of 0.15–3.5 kb. This gives an average of 8.18 bands per primer. Of the 90 bands, 53 proved to be polymorphic (56.17%) and hence informative (Table S2). RAPD-12 proved to be the most efficient with 81.81% of the 9 products being polymorphic. A representative RAPD profile obtained by primer RAPD-11 is shown in Fig. 1a. Of a total of 11 bands (0.15–3.5 kb), 6 are polymorphic (54.54%). Two distinct categories are visible. The first marked ‘M’ are monomorphic, and the second category marked ‘A’, a 1.2 kb band, is unique to genotypes from Africa (lanes 12–20, Fig. 1a), ‘B’, a 2.0 kb band, is unique to genotypes from Karnataka (lanes 1–4, Fig. 1a) and ‘C’, a 1.8 kb band, shared by accessions from Tamil nadu and Maharashtra (lanes 5–11, Fig. 1a). The PIC values, a reflection of allele diversity and frequency among the varieties, were not uniformly higher for all the RAPD loci tested. The PIC value ranged from 0.139(RAPD-21) to 0.487 (RAPD-011) with a mean of 0.280.
a RAPD profile of 83 finger millet genotypes generated by random primer (RAPD-11). The lanes represent, Lane L: 100 bp ladder; Lane 1–4 Karnataka, Lane 5–8 Uttar Pradesh, Lane 9–11 Tamil Nadu, Lane 12–20 African accessions and Lane M: λ-DNA marker. b SSR profile of 83 finger millet genotypes generated by primer (SSR-01). The lanes represent, Lane L: 100 bp ladder; Lane 1–4 Karnataka, Lane 5–8 Uttar Pradesh, Lane 9–11 Tamil Nadu, Lane 12–20 African accessions and Lane M: λ-DNA marker. c Cytochrome P450 gene based primer profile of 83 finger millet accessions generated by primer combination (Cyt 13(F) and cyt14(R). The lanes represent, Lane L: 100 bp ladder; Lane 1–4 Karnataka, Lane 5–8 Uttar Pradesh, Lane 9–11 Tamil Nadu, Lane 12–20 African accessions and Lane M: λ-DNA marker
SSR analysis
Ten SSR primers generated a total of 70 bands. Several polymorphic products specific to a particular geographic region could be observed. A representative fingerprint pattern generated by primer SSR-01 is shown in Fig. 1b. Out of 12 amplicons generated by this primer (size range 0.15–4.0 kb), eight were polymorphic. A 0.4 kb amplicon ‘A’ is present only in genotypes from Africa (lanes 12–20, Fig. 1b), and a 1.25 kb product is unique to genotypes from Karnataka (‘B’, lanes 1–4) and (‘C’, lanes 5–8, Fig. 1b) is shared by all genotypes except those from Uttar Pradesh (1.4 kb; Fig. 1b). Several other bands were shared by genotypes from most geographical regions. Such bands, exemplified by ‘D’, are shared by accessions from Tamil nadu and Maharashtra. The results are summarized in Table S3. As mentioned earlier, 5 SSR primers generated 70 bands (size range 0.15–4.0 kb), an average of 14 amplifications per primer. Fifty-one markers (70.19%) with an average of 10.2 per primer were polymorphic. PIC values for SSR primers were from 0.872 (SSR-02) to 0.922 (SSR-10) with an average 0.890.
Cytochrome P450 gene based markers analysis
All the 10 pairs of primers used in the present study were polymorphic, the number and size of polymorphic fragments revealed by each primer are presented in Table S4. A total of 103 scorable markers were yielded by the 10 pairs of polymorphic primers with an average of 10.3 bands per primer. Fifty-five markers (54.29%) with an average of 5.5 per primer were polymorphic. Cytochrome P450 gene based marker profile produced by the primer Cyt 13(F) and cyt14(R) is shown in Fig. 1c. Out of 11 amplified products, six are polymorphic. A (0.8 kb lanes 12–20, Fig 1c) and B (1.4 kb lanes 5–8, Fig 1c) are rare polymorphic bands as they are present in only genotypes from Africa and Uttar Pradesh, respectively. Another such product is indicated as ‘C’ which is absent from genotypes originating from Tamil Nadu and Karnataka. The PIC value ranged from 0.132 (Cyp2C19F and Heme2C19R) to 0.500 Cyt 015(F) and Cyt 016(R) with a mean of 0.327.
Statistical analysis
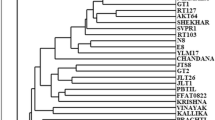
The binary matrix generated was used to calculate a similarity matrix based on Jaccard’s coefficient. The phenograms based on RAPD, SSR and cytochrome P450 gene based markers data indicate that accessions from different geographical regions are clearly distinguishable as separate clusters (Fig. S2a–c). All the phenograms confirm consistency of data. The principal correspondence analysis of RAPD, SSR and cyt P450 gene based markers (data not shown) reveal that all the genotypes belonging to a particular geographic region were grouped together in PCA plots except the genotypes from Uttar Pradesh as being most diverse. They are not only different from other genotypes but also from each other, and hence, cannot be grouped together. Mantel test [34] was employed to determine the coefficient of correlation between the similarity matrices generated by these markers. The coefficient of correlation between RAPD and SSR, RAPD and cyt P450 gene based primers and SSR and cyt P450 gene based primers was 0.675, 0.583 and 0.631, respectively. In E. coracana combined analysis of all the three markers (RAPD, SSR and cyt P450 gene based markers) grouped Uttar Pradesh genotypes into one tight cluster (Fig. S2d). Likewise, genotypes from Tamil Nadu and Karnataka formed distinct groups. But, genotypes from Africa appear diverse and showed relatedness to genotypes from Andhra Pradesh and Maharasthra. The coefficient of correlation between RAPD and combined RAPD, SSR and cyt P450 gene based primers, SSR and combined RAPD and SSR and cyt P450 gene based primers and cyt P450 gene based primers and combined RAPD and SSR and cyt P450 gene based primers was 0.607, 0.731 and 0.725, respectively. The three molecular marker systems were compared on the basis of different criteria (Table 1).
Dendrogram (unweighted pair group method analysis) clustered the finger millet accessions into two major groups, one comprised the accessions collected from north or northeast regions of India, and the other comprised five subclusters and the accessions collected from different states of southern India and Africa. In case of polymorphism detection, SSR as a tool scored higher than RAPD and cytochrome P450 gene based markers. Marker index ‘MI’ a measure of overall efficiency of a marker is highest for SSR, making it the most efficient marker for the genetic diversity studies in E. coracana. Mantel test was also employed to analyze the ‘goodness of fit’ for the UPGMA dendrograms generated by each marker system. This was done by making the cophenetic similarity matrices from each UPGMA phenogram and then comparing these cophenetic similarity matrices with the original similarity matrices (generated from binary data) for each marker technique. It revealed values higher than 0.90 for all the markers used [RAPD (r = 0.994, P = 0.01), SSR (r = 0.987, P = 0.01) and cytochrome P450 gene based markers (r = 0.940, P = 0.01)], thus confirming their authenticity and very good fit of clustering. Bootstrap analysis of this data was performed to determine the confidence values as percentages at each node. Very high bootstrap values were obtained from the major nodes.
Discussion
DNA markers have been used to evaluate genetic diversity in different crop species [37]. DNA fingerprinting is a routine method employed to study the extent of genetic diversity across a set of germplasm or cultivars and group them into specific categories. In general, random amplified polymorphic DNA (RAPD) is increasingly being employed in genetic research owing to its speedy process and simplicity [4]. In case of finger millet besides several phenotypic/morphological descriptors various molecular markers have been utilized for characterizing the germplasms. Based on seed colors as phenotypic descriptor, attempt has been made to analyze 36 genotypes of finger millet (Eleusine coracana (L.) Gaertn) and correlate with protein and calcium contents [38]. Several attempts have been made to assess the genetic diversity in finger millets using different DNA based molecular markers namely RAPD, RFLP and SSR. There are no reports of markers being used in characterization of accession of finger millet so far using markers based on Cyt P450 genes based markers. The rapidly evolving gene sequences of mixed function oxidases like cytochrome P450 can be exploited for assessing the intra-species diversity amongst finger millet genotypes.
Geographically isolated populations of plant species tend to accumulate genetic variations during the course of environmental adaptations. Plant species grow in greatly diverse environments. There is not only similarity, but also specificity or diversity as to the mechanisms employed by them in perceiving environment signals and adapting to the growth environments. In the present study distinct genetic identity present between genotypes was highlighted using DNA amplification profile procured from separate geographical regions similarly genetic divergence and phylogenetic analysis of genus Jatropha widely distributed in India has been done based on nuclear ribosomal DNA ITS sequence [39].
All the molecular markers, used in the present study detected polymorphism percentage ranging between 20 and 80%. RAPD revealed an average of 56.17% polymorphism across all accessions. Fakrudin et al. [24] also reported 85.82% of polymorphism in 12 finger millet accessions with RAPDs. Such high percentage polymorphism (91%) is also reported in 32 accessions of E. coracana by Babu et al. [25]. SSR revealed an average of 70.19% polymorphism across all accessions. Similar results were also reported by Dida et al. [28] in E. coracana. Cyt P450 genes based markers revealed an average of 54.29% polymorphism across all accessions. Among the three marker systems employed, 18 RAPD primers produced a total of 90 markers whereas SSR and 10 pairs of cytochrome P450 gene based primers produced 70 and 103 markers, respectively. The level of polymorphism revealed by SSR (70.19%) is higher than RAPD (56.17%) followed by cytochrome P450 gene based markers (54.29%).
In the present investigation, the average number of fragments amplified by RAPD primers among the 83 accessions was 8.18 with a range of 1–15. Fakrudin et al. [24] reported an average of 6 markers amplified by each primer in Eleusine coracana. Such a high variation in the number of fragments produced by these arbitrary primers may be attributed to the differences in the binding sites throughout genome of the genotypes included. Cytochrome P450 gene based primers generated 2–13 markers with average of 10.3 per genotype. Earlier, generation of 7 markers in Cymbopogon species by cytochrome P450 gene based primers were reported by Kumar et al. [40].
Comparison of PIC values for three marker systems (a parameter associated with the discriminating power of markers) indicated that the range of PIC values for RAPD primers was from 0.139(RAPD-21) to 0.487 (RAPD-011) with a mean of 0.280. PIC values for SSR primers were from 0.872 (SSR-02) to 0.922 (SSR-10) with an average 0.890. The comparison of the average PIC values of cytochrome P450 gene based primers revealed that the lowest was with 0.132 (Cyp2C19F and Heme2C19R) and the highest was with 0.500 Cyt 015(F) and Cyt 016(R) with a mean of 0.327.
The coefficient of correlation between RAPD and SSR, RAPD and cyt P450 gene based primers was 0.675, 0.583 and 0.631, respectively. RAPD and combined RAPD,SSR and cyt P450 gene based primers, SSR and combined RAPD and SSR and cyt P450 gene based primers and cyt P450 gene based primers and combined RAPD and SSR and cyt P450 gene based primers was established by means of high value of matrix correlation value of 0.607, 0.731 and 0.725, respectively. Similar results have been obtained with RAPD and ISSR in Vigna umbellate by Muthusamy et al. [41]. Hence, all the marker systems RAPD, SSR and cytochrome P450 gene based markers either individually or combined can be effectively used in determination of genetic relationships among finger millet genotypes.
Cluster analysis was carried out on four sets of marker profiling data based on (1) RAPD, (2) SSR, (3) cyt P450 gene based primers, (4) combination of RAPD, SSR and cyt P450 gene based primers. Dendrogram clustered the finger millet accessions into two major groups, one comprised the accessions collected from north or northeast regions of India, and the other comprised five subclusters and the accessions collected from different states of southern India and Africa. In E. coracana individual and combined analysis of all the three markers grouped Uttar Pradesh genotypes into one tight cluster. Likewise, genotypes from Tamil Nadu and Karnataka formed another two distinct groups. But genotypes from Africa appear diverse but showed relatedness to genotypes from Andhra Pradesh and Maharashtra. Distinct clusters were formed by the genotypes of southern India and northern India. The bootstrap values for the phenogram generated from combination of RAPD, SSR and cyt P450 gene based markers are 100% for the clusters (Africa, Tamil Nadu and Andhra Pradesh). Das et al. [42] detected highest similarity coefficient in the genotypes collected from southern India and the least from northern India. The results of present investigation also suggest the close relationship within the genotypes collected from different states of southern India as they are showing highest similarity coefficient and clustered together along with African genotypes. Several reasons could be attributed to the low intra-zonal diversity detected in the present investigation. First, the genotypes have genetically adapted to local environment and have become homogenous as a result of ‘inbreeding’ or exchange of genetic material within the said population. Thus, in the present investigations, unique DNA amplification profile brought out a distinct genetic identity belonging to genotypes of separate geographical regions.
In the present investigation, various high intensity bands were amplified, like a 0.4 kb band amplified by primer SSR-01, a 1.2 kb band amplified by primer RAPD-011 and a 0.8 kb band amplified by primer (Cyt 13(F) and cyt 14(R) only in African accessions. The region-specific bands detected in our studies can be used for the authentication of this crop. Das et al. [42] have already exploited RAPD for confirming the genuineness of E. coracana. Salimath et al. [23] have detected ISSR and RAPD primers which can discriminate between accessions of E. coracana from Africa and Asia. Microsatellites detect high level of polymorphism, which can be used to discriminate between closely related genotypes [43].
The comparison of the three molecular marker systems revealed SSR to be the best as it generated highest percentage polymorphism and high MI. Phenograms generated from RAPD, SSR and cytochrome P450 gene based markers data showed similarity in relative placement of genotypes. SSR and cytochrome P450 gene based markers provided a more detailed idea on the genetic relationship.
The cophenetic correlation values of Mantel test are above 0.90 for each marker system. This validates the phenograms obtained after UPGMA analysis of binary matrices data. These findings can be used in authentication of the nutritionally important crop and perhaps in recognizing high yielding population in a particular area. Exploration and evaluation of diversity among these germplasm would be of great significance for in situ conservation and finger millet breeding programmes.
Abbreviations
- PCA:
-
Principal component analysis
- PCR:
-
Polymerase chain reaction
- PIC:
-
Polymorphism information content
- RAPD:
-
Random amplified polymorphic DNA
- UPGMA:
-
Unweighted pair group method with arithmetic averages
References
Venkannababu B, Ramana T, Radhakrisnan TM (1987) Chemical composition and protein content in hybrid varieties of finger millet. Indian J Agric Sci 57:520–522
Chennaveeraiah MS, Hiremath SC (1991) Cytogenetics of minor millets. In: Tsuchiya T, Gupta PK (eds) Chromosome engineering in plants genetics breeding and evolution. Elsevier, Amsterdam, pp 613–627
Chethan S, Malleshi NG (2007) Finger millet polyphenols: characterization and their nutraceutical potential. Am J Food Technol 27:282–292
Williams JGK, Kubelik AR, Livak KJ, Rafalski AJA, Tingey SV (1990) DNA polymorphism amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 22:6531–6535
Jacobson A, Hedrén M (2007) Phylogenetic relationships in Alisma Alismataceae based on RAPDs, and sequence data from ITS and trnL. Plant Syst Evol 265:27–44
Soller M, Beckmann JS (1983) Genetic polymorphism in varietal identification and genetic improvement. Theor Appl Genet 67:25–33
Vos PR, Hogers R, Bleeker M, Reijans M, Lee T, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for fingerprinting. Nucleic Acids Res 23(21):4407–4414
Zietkiewicz E, Rafalski A, Labuda D (1994) Genome fingerprinting by simple sequence repeat SSR-anchored polymerase chain reaction amplification. Genomics 202:176–183
Becker J, Heun M (1994) Barley microsatellites: allele variation and mapping. Plant Mol Biol 274:835–845
Garcia AAF, Benchimol LL, Barbosa AMM, Geraldi IO, Souza CLJ, Souza AP (2004) Comparison of RAPD, RFLP, AFLP and SSR markers for diversity studies in tropical maize inbred lines. Genet Mol Biol 27:579–588
Nazari L, Pakniyat H (2008) Genetic diversity of wild and cultivated Barley genotypes under drought stress using RAPD markers. Biotechnology 74:745–750
Malik AR, Pervaiz ZH, Masood MS (2008) Genetic diversity analysis of traditional and improved cultivars of Pakistani rice Oryza sativa L. using RAPD markers. Electron J Biotechnol 11(3):1–10
Davierwal AAP, Chowdari KV, Shiv K, Reddy APK, Ranjekar PK, Gupta VS (2000) Use of three different marker systems to estimate genetic diversity of Indian elite rice varieties. Genetica 108:269–284
Rashed MA, Abou-Deif MH, Sallam MAA, Ramadan WA (2008) Estimation of genetic diversity among thirty bread wheat varieties by RAPD analysis. J Appl Sci Res 412:1898–1905
Colombo C, Second G, Valle TL, Charrier A (1998) Genetic diversity characterization of cassava cultivars Manihot esculenta Crantz. I. RAPD markers. Genet Mol Biol 21:69–84
Chao S, Zhang W, Dubcovsky J, Sorrells M (2007) Evaluation of genetic diversity and genome wide linkage is equilibrium among US wheat triticum aestivum L germplasm representing different market classes. Crop Sci 47:1018–1030
Agrama HA, Tuinstra MR (2003) Phylogenetic diversity and relationships among sorghum accessions using SSRs and RAPDs. Afr J Biotechnol 2:334–340
Qi-lun Y, Ping F, Ke-cheng K, Guang-Tang P (2008) Genetic diversity based on SSR markers in maize Zea mays L. landraces from Wuling mountain region in China. J Genet 87:287–291
Shalk M, Nedelkina S, Schoch G, Batard Y, Werck-reichhart D (1999) Role of unusual amino-acid residues in the proximal and distal heme regions of a plant P450, CYP73A1. Biochemistry 38:6093–6103
Tanksley SD, Mccouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
Somerville C, Somerville S (1999) Plant functional genomics. Science 285:380–383
Yamanaka S, Suzuki E, Tanaka M, Takeda Y, Watanabe JA, Watanabe KN (2003) Assessment of cytochrome P450 sequences offers a useful tool for determining genetic diversity in higher plant species. Theor Appl Genet 108:1–9
Salimath SS, Olivera ACD, Godwin ID, Bennetzen JL (1995) Assessment of genome origins and diversity in the genus Eleusine with DNA markers. Genome 38:757–763
Fakrudin B, Kulkarni RS, Shashidhar HE, Hittalmani S (2007) Genetic diversity assessment of finger millet, Eleusine coracana, germplasm through RAPD analysis. PGR Newslett 138:52–54
Babu B, Senthil N, Gomez S, Biji K, Rajendraprasad N, Kumar S, Babu R (2007) Assessment of genetic diversity among finger millet Eleusine coracana L Gaertn accessions using molecular markers. Genet Resour Crop Evol 54:399–404
Parani M, Rajesh K, Lakshmi M, Parducci L, Szmidt AE, Parida A (2001) Species identification in seven small milletspecies using polymerase chain reaction–restriction fragment length polymorphism oftrnS-psbC gene region. Genome 44:495–499
Hilu KW (1995) Evolution of finger millet, evidence from random amplified polymorphic DNA. Genome 38:232–238
Dida MM, Wanyera N, Dunn MLH, Bennetzen JL, Devos KM (2008) Population structure and diversity in finger millet Eleusine coracana germplasm. Trop Plant Biol 1:131–141
Dida MM, Srinivasachary, Ramakrishnan S, Bennetzen JL, Gale MD, Devos KM (2007) The genetic map of finger millet, Eleusine coracana. Theor Appl Genet 114:321–332
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4326
Maniatis T, Sambrook J, Fritsch EF (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Rohlf FJ (2001) NTSYS-pc numerical taxonomy and multivariate analysis system. Version 5.1. Exeter Publishing Ltd., Setauket
Jaccard P (1908) Nouvelles recherches sur la distribuition florale. Bull Soc Vaud Sci Nat 44:223–270
Mental N (1967) The detection of disease clustering and generalized regression approach. Cancer Res 27:209–220
Yap IV, Nelson R (1995) WinBoot: a program for performing bootstrap analysis of binary data to determine the confidence limits of UPGMA-based dendrograms. IRRI discussion paper series no. 14. IRRI, Los Banos
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR microsatellite marker for germplasm analysis. Mol Breed 2:225–238
Cooke RJ (1995) Variety identification of crop plants. In: Skerrit JH, Appels R (eds) New diagnostics in crop science. Biotechnology in Agriculture No. 13 CAB International, Wallingford, UK, pp 33–63
Vadivoo AS, Joseph R, Ganesan NM (1998) Genetic variability and diversity for protein and calcium contents in finger millet Eleusine coracana L. Gaertn in relation to grain color. Plant Foods Hum Nutr 52:353–364
Pamidimarri DVNS, Chattopadhyay B, Reddy MP (2009) Genetic divergence and phylogenetic analysis of genus Jatropha based on nuclear ribosomal DNA ITS sequence. Mol Biol Rep 36:1929–1935
Kumar J, Verma V, Qazi GN, Gupta PK (2007) Genetic diversity in cymbopogon species using PCR-based functional markers. J Plant Biochem Biotechnol 16:119–122
Muthusamy S, Kanagarajan S, Ponnusamy S (2008) Efficiency of RAPD and ISSR markers system in accessing genetic variation of rice bean Vigna umbellata landraces. Electron J Biotechnol 11(3):1–10
Das S, Mishra RC, Rout GR, Aparajita S (2007) Genetic variability and relationships among thirty genotypes of finger millet Eleusine coracana L. Gaertn. using RAPD markers. J Biosci 621–2:116–122
Teulat B, Aldam C, Trehin R, Lepbrun P, Barker JHA, Arnold GM, Karp A, Baudouin L, Rognan F (2000) An analysis of genetic diversity in coconut Cocos nucifera populations from across the geographic range using sequence tagged microsatellites SSRs and AFLPs. Theor Appl Genet 100:764–771
Acknowledgments
The authors wish to acknowledge the Department of Biotechnology, Govt. of India for providing financial support in the form of Programme Support for research and development in Agricultural Biotechnology at G.B. Pant University of Agriculture and Technology, Pantnagar (Grant No. BT/PR7849/AGR/02/374/2006). The first author was supported by fellowship from DBT, India during the project programme. The authors thank the All India Co-ordinated Small Millets Improvement Project, ICAR, UAS, GKVK, Bangalore for providing the seed samples of germplasm analysed in present study. We are highly thankful to Dr. T. Mohapatra for his kind academic inputs in data analysis and interpretation of data.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Panwar, P., Saini, R.K., Sharma, N. et al. Efficiency of RAPD, SSR and Cytochrome P450 gene based markers in accessing genetic variability amongst finger millet (Eleusine coracana) accessions. Mol Biol Rep 37, 4075–4082 (2010). https://doi.org/10.1007/s11033-010-0067-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0067-5