Abstract
Bim is a proapoptotic member of the Bcl-2 family and is primarily involved in the regulation of the intrinsic apoptotic pathway. However, the detail of regulation of Bim’s proapoptotic activity has not been clarified yet. Using Bim L as bait, we screened a human fetal cDNA library for interacting proteins and identified Grb10 as an interactor. This interaction was verified by co-immunoprecipitation and intracellular co-localization studies. The potential segment of Bim L that binds Grb10 was identified via a yeast mating test. Grb10 interacted with the DBD (dynein binding domain) of Bim and inhibited apoptosis triggered by overexpression of DBD containing Bim isoforms. The putative phosphorylation sites on DBD of Bim play a role for the anti-proapoptotic activity of Grb10. Our results suggest that Grb10 interacts with Bim L and inhibits its proapoptotic activity in a phosphorylation-dependant manner.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Apoptosis is a physiological progress for killing unwanted cells in multicellular organisms that is necessary for embryonic development and tissue homeostasis [1]. There are two main intracellular apoptosis cascades—one that involves the interaction of a death receptor with its ligand and another with involves the mitochondria. The mitochondrial pathway is regulated by proapoptotic and antiapoptotic Bcl-2 family proteins [2]. The antiapoptotic proteins, such as Bcl-2, Bcl-xL, Mcl-1 and Boo (Diva), contain at least a BH1 and BH2 domain. The proapoptotic members consist of two subgroups: Bax, Bak and Bok (Mtd), which share homology in the BH1, BH2 and BH3, but not in BH4 domains; Bim, Bid, Bad, Bik, Bmf, Hrk, Noxa and Puma, which share homology only in the BH3 domain [3].
The BH3-only protein Bim was first discovered by an expression screen for proteins that bind to Bcl-2 [4]. To date, there are at least 15 isoforms of Bim that have been reported [5, 6]. All three major isoforms of Bim, Bim EL, L and S, are able to bind to Bcl-2 and Bcl-xL, which reside on the mitochondrial outer membrane, and initiate cytochrome C release in certain settings of apoptosis. The apoptotic activity of Bim EL and Bim L is suppressed by binding to the dynein motor complex via the dynein binding domain (DBD). Bim S lacks the DBD and thus has most potent cytotoxic activity [7]. At present, the studies of transcriptional and posttranslational regulation of Bim have revealed that the transcriptionof Bim was regulated by HDACi and FOXO3a, while the modification of Bim is effected by kinase family members such as JNK and ERK [8–10]. However, the upstream regulators and downstream targets of Bim remain largely unknown [11, 12].
Here we report that Bim L interacts with Grb10, an adapter protein that is known to interact with a number of receptor tyrosine kinases and signaling molecules. The function of this interaction was determined by apoptosis assays.
Materials and methods
Plasmid construction
The pLexA-Bim bait plasmids for the yeast two hybrid assay and pEGFP-Bim plasmids for expressing EGFP-Bim fusion proteins were described previously [6].
The ORF of human Grb10 was obtained from a human fetal cDNA library (Clontech) by amplification with the following primers: 5′-ccggaattctgcaagctgccggccctctgttc-3′; 5′-cgcggatccaaggccactcggatgcagtggtg-3′. The cDNA was inserted into pDsRed-N1 (Clontech) between the EcoRI and BamHI sites to generate pGrb10-DsRed that expresses a Grb10-DsRed fusion protein. The Grb10 SH2 domain fragment was amplified from a pB42AD fetal brain cDNA library (Clontech) with the pB42AD sequence primers, 5′-ccagcctcttgctgagtggagatg-3′ and 5′-ggagacttgaccaaacctctggcg-3′, and subcloned into pcDNA3.0 (Clontech) between the EcoRI and Hind III sites for eukaryotic expression of the protein.
To identify the effect of mutation of the phosphorylation sites of Bim L on the Bim L and Grb10 interaction, the plasmids that express Bim L-GFP with mutations in putative phosphorylation sites were generated. Using Net phos2.0 (http://www.cbs.dtu.dk/services/NetPhos/) and the GPS Group-based Phosphorylation Scoring Method (url Version 1.10 (http://973-proteinweb.ustc.edu.cn/gps/gps_web/predict.php) [13, 14], the phosphorylation sites in Bim L were predicted. Bim LY102Q, Bim LY103F and Bim LY102QY103F are mutations of the putative phosphorylation sites near the BH3 domain, and Bim LS44A and Bim LT56A are mutations of the putative phosphorylation sites in the DBD. All site-directed mutagenesis was performed using the Muta-direct kit (SBS, China). The primers used for the mutations mentioned above were 5′-gacgagtttaacgctcaatatgcaaggaggg-3′ and 5′-ccctccttgcatattgagcgttaaactcgtc-3′, 5′-cgagtttaacgcttactttgcaaggagggtatttttg-3′ and 5′-caaaaataccctccttgcaaagtaagcgttaaactcg-3′, 5′-cgagtttaacgctcaatttgcaaggaggg-3′ and 5′-ccctccttgcaaattgagcgttaaactcg-3′, 5′-cagagccacaagacagggccccagcacccatgagttg-3′ and 5′-caactcatgggtgctggggccctgtcttgtggctctg-3′ and 5′-gttgtgacaaatcaacacaagccccaagtcctccttgccag-3′ and 5′-ctggcaaggaggacttggggcttgtgttgatttgtcacaac-3′, respectively.
Yeast two-hybrid assay
The pLexA two-hybrid system was purchased from Clontech. Using pLexA-Bim L as bait, the yeast two-hybrid was performed using a standard two-step protocol to screen a pre-made pB42AD-fetal cDNA library (Clontech). All positive clones were amplified with the pB42AD sequence primers (5′-ccagcctcttgctgagtggagatg-3′ and 5′-ggagacttgaccaaacctctggcg-3′) and PCR products were sequenced with the same primers. The yeast-mating test was also used to determine the interaction between the other three Bim isoforms with the sequenced pB42AD positive clones. pLexA-Bim S, pLexA-Bim α2, pLexA-Bim α3 was used to transfect YM4271, and was then mated with the pB42AD-positive clone EGY48 [p8opLacZ] yeast cells.
Co-immuprecipitation
To verify the interaction between Bim L and Grb10, co-immuprecipitation was performed. HEK293 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal calf serum, 50 μg streptomycin/ml and 50 IU penicillin/ml. The cultures were incubated at 37°C in a humidified atmosphere of 5% CO2. The cells were transfected with plasmid pEGFP- Bim L or control plasmid pEGFP-C1 using Lipofectamine (Invitrogen). 24 h after transfection, the cells were harvested and lysed in 500 μl lysis buffer containing 20 nM Tris pH 7.5, 100 mM NaCl, 0.5% NP-40, 0.5 mM EDTA, 0.5 mM PMSF and 0.5% protease inhibitor cocktail (Sigma). Five micrograms of rabbit anti-Grb10 monoclonal antibody (Santa Cruz) was added to 500 μl of each cell lysate. After a 2-h incubation of the immunoprecipitates at 4°C, 10 μl of pre-washed protein A/G Sepharose (Santa Cruz) were added to each immunoprecipitated complex and shaken for 2 h at 4°C. The immunoprecipitates were washed three times with ice-cold wash buffer. Lysates and washed immunoprecipitates were resolved on SDS-PAGE and transferred to nitrocellulose for western blot assays. The nitrocellulose membrane was blocked with 5% nonfat milk and incubated overnight at 4°C in mouse anti-EGFP antibody (Santa Cruz). Then the membrane was exposed to horseradish-peroxidase conjugated anti- mouse antibody (Santa Cruz) and the bands were visualized using an enhanced chemiluminescence immunoblotting detection kit (Amersham Biosciences).
Subcellular localization
The subcellular localization of Bim L and Grb10 were determined in HEK293 cells. HEK293 cells were plated at a density of 2 × 105 cells per well (6-well plate). When the cells were about 75% confluent, cells were transfected with pEGFP-Bim L and pGrb10-DsRed using Lipofectamine (Invitrogen), and the pDsRed and pEGFP-C1 plasmids were co-transfected as controls with the above two plasmids, respectively. Twenty-four hours after transfection, the cells were observed using a confocal fluorescence microscope (Leica). The nuclei were strained with DAPi. The fluorescence of GFP, RFP and DAPi was observed at wavelengths of 488, 543 and 405 for excitation and 510, 582 and 461 for emission, respectively.
Apoptosis assay
HEK293 cells were plated at a density of 2 × 105 cells per well. When the plates were about 75% confluent, cells were co-transfected with pEGFP-Bim isoforms/mutatants and pcDNA3.0-Grb10SH2-BPS using the Lipofectamine agent (Invitrogen). pEGFP-Bim isoforms/mutatants and pcDNA3.0 co-transfected cells were used as a control. Twenty-four hours after transfection, the cells were harvested. Apoptotic cells were quantified by flow cytometric analysis (APO-BRDUTM Apoptosis Assay Kit, Becton Dickinson) according to the manufacturer’s recommendations. The experiment was repeated three times.
Results
Yeast two-hybrid library screening with Bim L
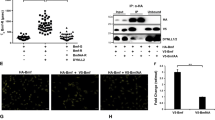
pLexA-Bim L was used as bait to screen a pB42AD-based human fetal brain cDNA library for potential Bim L-interacting proteins. A total of 121 positive clones were isolated from 106 independently screened clones. Of the 121 positive clones, 14 corresponded to the Grb10 C-terminus, which encoded part of the BPS domain and the complete SH2 domain. In addition, the mating test revealed that four isoforms of Bim L (Bim S, Bim α2, Bim α3,) interact with Grb10 with different interaction strengths, the interaction between Bim L and Bim α2 are strenger then that of Bim S and Bim α3 (Fig. 1).
Co-immuprecipitation indicated an interaction between Bim L and Grb10
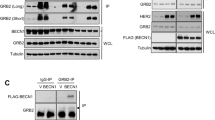
To further investigate the interaction between Bim L and Grb10 in vivo, co-immuprecipitation was performed in HEK293 cells overexpressing the EGFP-Bim L fusion protein. A rabbit anti-Grb10 antibody was able to precipitate EGFP-Bim L from the lysate of HEK293 cells transfected with pEGFP-Bim L, while there was no band present in the control co-immuprecipitation of pEGFP-C1 transfected cells (Fig. 2).
Co-immunoprecipitation analysis of Grb10 and Bim L in HEK293 cells. Samples were analyzed by western blotting with a mouse anti-GFP antibody. 1 Cell lysate of pEGFP-C1-transfected HEK293 cells were incubated with a rabbit anti-Grb10 binding protein on A/G agarose beads. 2 Cell lysate of pEGFP-Bim L-transfected HEK293 cells were incubated with a rabbit anti-Grb10 binding protein on A/G agarose beads. 3 Cell lysate of pEGFP-C1-transfected HEK293 cells. 4 Cell lysate of pEGFP-Bim L-transfected HEK293 cells
Intracellular localization of Bim L and Grb10
In order to investigate the intracellular localization of Bim L and Grb10, HEK293 cells were co-transfected with pEGFP-Bim L and Grb10-DsRed plasmids. Both the EGFP-Bim L and Grb10-DsRed fusion proteins were detected in the cytoplasm, and especially in irregular particles surrounding the nucleus. In GFP-Bim L and Grb10-DsRed-expressing cells, the green fluorescence and red fluorescence merged to some degree (Fig. 3).
Grb10 inhibited Bim L induced apoptosis
To investigate the function of Grb10 in apoptosis induced by Bim, the apoptosis in pEGFP-Bim and pcDNA3.0-Grb10SH2-BPS co-expression HEK293 cells were assayed. Compared with the control, in Grb10 over-expressing HEK293 cells, the apoptosis induced by over-expression of Bim L is significantly decreased (from 69.66 ± 1.31 to 48.34 ± 2.04, n = 3, P < 0.005). The results of the apoptosis assay showed that Grb10SH2-BPS apparently inhibits apoptosis induced by pEGFP-Bim α2 (from 31.78 ± 1.55 to 17.36 ± 0.89, n = 3, P < 0.005), pEGFP-Bim LY102Q (from 66.13 ± 1.65 to 47.26 ± 2.55, n = 3, P < 0.005) and pEGFP-Bim LY103F (from 64.65 ± 1.99 to 47.62 ± 1.80, n = 3, P < 0.005). Apoptosis of cells co-transfected with Grb10SH2-BPS and pEGFP-Bim S (from 68.71 ± 1.92 to 67.66 ± 1.45, n = 3, P < 0.05), pEGFP-Bim α3 (from 35.48 ± 1.35 to 34.83 ± 2.03, n = 3, P < 0.05), pEGFP-Bim LS44A (from 72.35 ± 1.87 to 69.88 ± 1.74, n = 3, P < 0.05) and pEGFP-Bim LT56A (from 69.53 ± 1.26 to 67.52 ± 1.12, n = 3, P < 0.05) did not presented significant difference (Fig 4).
Discussion
Grb10 was isolated as a Bim interactor by yeast-two-hybridization. The interaction between Bim and Grb10 was further confirmed by co-immunoprecipitation. The localization of Bim L-EGFP and Grb10-DsRed overlapped somewhat, which supports the possibility of an interaction between Bim L and Grb10. Through apoptosis assays, the function of the interaction between these two proteins was also explored. Over-expression of Grb10 apparently inhibited Bim L or Bim α2-induced apoptosis. However, over-expression of Grb10 was not able to inhibit apoptosis induced by Bim S, or Bim α3, which lacks the DBD domain. The yeast-mating test showed that Bim isoforms containing the DBD domain interacted with Grb10 stronger then the Bim isoforms without DBD. The results supported that DBD play a role in Grb10 mediated anti-apoptotic activity.
The phosphorylation state of Bim L is complex and plays an important role in its apoptotic activity [9]. We analyzed apoptosis induced by Bim L phosphorylation mutants (BimS44A, Bim LT56A, Bim LY102Q, Bim LY103F and Bim LY102QY103F) in Grb10 co-expressing HEK293 cells. Apoptosis induced by mutants that changed the DBD domain phosphorylation sites (Bim LS44A and Bim LT56A) could not be inhibited by over-expression of Grb10. Grb10 belongs to a small family of adapter proteins that are known to interact with a number of receptor tyrosine kinases and signaling molecules. It mediated the phosphorylation protein degeneration by targeting them for ubiquitination [15–17]. JNK phosphorylation on Thr56 and Ser44 released Bim L from the dynein motor complex and increased its apoptotic activity [9]. The Ser44 in Bim L has also been found to be phosphorylated by ERK1/2 [18]. Our results suggested that the phosphorylation sites of DBD domain are crucial for Grb10 anti-apoptotic activity.
Our study first revealed that Grb10 can interact with a proapoptotic protein, Bim. It is notable that the region of Bim L interacting with Grb10 is not the BH3 domain, but rather the DBD domain. The anti-apoptotic activity of Grb10 is depended on the phosphorylation of DBD of Bim. Our research shed a light on the Bim proapoptotic activity regulation by Grb10 and the phosphorylation. However, the meaning of interaction between Bim and Grb10 need further study.
References
Jacobsen MD, Weil M, Raff MC (1997) Programmed cell death in animal development. Cell 88:347–354
Adams JM, Cory S (1998) The Bcl-2 protein family: arbiters of cell survival. Science 281:1322–1326
Cory S, Huang DC, Adams JM (2003) The Bcl-2 family: roles in cell survival and oncogenesis. Oncogene 22:8590–8607
O’Connor L, Strasser A, O’Reilly LA, Hausmann GM, Adams J, Cory S, Huang DC (1998) Bim: a novel member of the Bcl-2 family that promotes apoptosis. EMBO J 17:384–395
Mami U, Miyashita T, Shikama Y, Tadokoro K, Yamada M (2001) Molecular cloning and characterization of six novel isoforms of human Bim, a member of the proapoptotic Bcl-2 family. FEBS Lett 509:135–141
Chen JZ, Ji CN, Gu SH, Li JX, Zhao EP, Huang Y, Huang L, Ying K, Xie Y, Mao YM (2004) Over-expression of Bim α3, a novel isoform of human Bim, result in cell apoptosis. Int J Biochem Cell Biol 36:1554–1561
Puthalakath H, Huang DC, O’ Reilly LA, King SM, Strasser A (1999) The proapoptotic activity of the Bcl-2 family member Bim is regulated by interaction with the dynein motor complex. Mol Cell 3:287–296
Harris CA, Johnson EM Jr (2001) BH3-only Bcl-2 family members are coordinately regulated by the JNK pathway and require Bax to induce apoptosis in neurons. J Biol Chem 276:37754–37760
Lei K, Davis RJ (2003) JNK phosphorylation of Bim-related members of the Bcl2 family induces Bax-dependent apoptosis. PNAS 100:2432–2437
Putcha GV, Le S, Frank S, Besirli CG, Clark K, Chu B, Alix S, Youle RJ, LaMarche A, Maroney AC, Johnson EM Jr (2003) JNK-mediated BIM phosphorylation potentiates BAX-dependent apoptosis. Neuron 38:899–914
Adachi M, Zhao X, Imai K (2005) Nomenclutrue of dynein light chain-linked BH3-only protein Bim isoforms. Cell Death Differ 12:192–193
Pinon JD, Labi V, Egle A, Villunger A (2009) Bim and Bmf tissue homeostasis and malignant disease. Oncogene 27:s41–s52
Wan J, Kang SL, Tang C, Yan JH, Ren YL, Liu J, Gao XL, Banerjee A, Ellis LBM, Li TB (2008) Meta- prediction of phosphorylation sites with weighted voting and restricted grid search parameter selection. Nucleic Acids Res 36:1–11
Xue Y, Zhou F, Zhu M, Ahmed K, Chen G, Yao X (2005) GPS: a comprehensive www server for phosphorylation sites prediction. Nucleic Acids Res 33:W184–W187
Riedel H (2004) Grb10 exceeding the boundaries of a common signaling adapter. Front Biosci 9:603–618
Morrione A (2003) Grb10 adapter protein as regulator of insulin-like growth factor receptor signaling. J Cell Physiol 197:307–311
Morrione A, Plant P, Valentinis B, Staub O, Kumar S, Rotin D, Baserga R (1999) mGrb10 interacts with Nedd4. J Biol Chem 274:24094–24099
Ley R, Ewings KE, Hadfield K, Cook SJ (2005) Regulatory phosphorylation of Bim: sorting out the ERK from the JNK. Cell Death Differ 12:1008–1414
Acknowledgements
The authors acknowledge the support of the State Hightech Development Program (2007AA021002) the Nature Science Fund (Grant No. 30771236).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Authors Zhi-qian Hu and Jia-yi Zhang contributed equally.
Rights and permissions
About this article
Cite this article
Hu, Zq., Zhang, Jy., Ji, Cn. et al. Grb10 interacts with Bim L and inhibits apoptosis. Mol Biol Rep 37, 3547–3552 (2010). https://doi.org/10.1007/s11033-010-0002-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-010-0002-9