Abstract
Glutamate decarboxylase produces GABA, the main inhibitory neurotransmitter in adult mammalian brain. Two homologous forms of GAD encoded by separate genes have been identified in mammalian brain, with molecular weight of 67 kDa (GAD67) and 65 kDa (GAD65). Here, we studied the transcriptional regulation of GAD67. Three transcript variants (GAD67A, GAD67B, and GAD67C) transcribed from distinct categories of transcriptional start sites were identified. RT-PCR revealed these transcripts have distinct tissues distributions. Though GAD67A and GAD67B were co-expressed in brain and many nonneural tissues, in heart, only GAD67A was expressed. GAD67C was specifically expressed in testis. These transcripts also showed distinct developmental expression patterns during testis maturation. GAD67A was expressed at all age points examined. GAD67B was only detected at postnatal day 1 and day 5, while GAD67C was expressed from postnatal day 30. Characterizing the genome sequence upstream of transcriptional start sites of these transcripts revealed the presence of TATA-less promoters. Potential promoter activities were analyzed by coupling these promoter sequences to the open reading frame of a luciferase reporter gene in transient expression experiments. Moreover, our results showed GAD67 gene expression was also regulated by alternative splicing in postnatal day 1 and day 5 testis. The above results suggested GAD67 gene expression was dynamically regulated by alternative promoters and splicing during postnatal rat testis maturation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
GABA is the most important inhibitory neurotransmitter in central nervous system. It was also present in the mammalian testes, where may serve as a paracrine modulator of cell proliferation, migration, and differentiation [1]. Glutamate decarboxylase [2] is the key enzyme that catalyzes the formation of the GABA from glutamate acid. Encoded by separate genes, two isoforms of GAD, namely, a 65 kDa (GAD65) and a 67 kDa (GAD67), are found in mammalian CNS. The two GADs differ in sequence, subcellular distribution, and interactions with pyridoxal phosphate, their obligate cofactor. GAD is expressed primarily in GABAergic neurons of the adult mammalian central nervous system, but it is also present in pancreatic β-cells and testis [3, 4]. The change of GABA level in CNS has been related to several neuropathological disorders, such as Alzheimer’s disease, Parkinson’s disease, and seizure [5–7]. Autoantibodies to glutamic acid decarboxylase (GAD) are frequent at or before the onset of insulin-dependent diabetes mellitus [8]. Thus study of GAD gene expression is of significant value for understanding the mechanism of these diseases.
Previous research proved GAD67 expression can be regulated by multiple mechanisms [9, 10]. GAD67 gene expression was developmentally regulated by alternative splicing in embryonic rodent brains [11, 12]. The alternative splicing resulted in two additional GAD67 transcripts in which either the first 80 or the first 86 bp of exon 7 were inserted into the full length GAD67 mRNA, respectively. Transcripts with the 80-bp insertion result in two overlapping open reading frames, encoding an enzymatically inactive 25-kDa protein (GAD25) and an enzymatically active 44-kDa protein (GAD44). A second stop codon at the end of the 86-bp insertion abolishes the translation of GAD44 [11]. Recent genome wide analysis revealed many genes possess more than one promoter, and alternative promoters play an important role in regulating gene expression [13, 14]. For rat GAD65 gene, multiple transcription start sites (TSSs) transcribed from two independent TATA-less promoters was reported, and the two alternative promoters were developmentally used during testis maturation. Mouse GAD67 gene possesses a proximal housekeeping promoter and two more distal, conventional (TATA) promoters (AF354680). However, for rat GAD67 gene, so far, only one TATA-less promoter was identified [15], whether rat GAD67 gene also contain multiple promoters and play a role in regulating gene expression was not known.
The aim of the present study was to elucidate the molecular mechanism that directs the expression of GAD67 gene. Three distinct GAD67 transcripts, distinguished by their different promoter usage and alternative splicing, were identified. These transcripts have distinct tissues distributions, and showed age-dependent expression patterns during testis maturation. The promoter activity of the 5′-flanking genomic regions of each transcript in rat cell lines was further confirmed by using a transient expression system. Our results suggested GAD67 gene expression was dynamically regulated by alternative promoters and splicing during postnatal rat testis maturation.
Materials and methods
Animal
Sprague–Dawley rats were obtained from Laboratory Animal Center (Shanghai, China). All animal studies were carried out according to local and national guidelines for the care and use of laboratory animals and approved by Biomedical Research Ethics Committee of Shanghai Institute of Biological Sciences.
RACE-PCR
The RACE experiments were performed using the Smart RACE cDNA Amplification Kit (Clontech). The 5′ RACE-ready cDNAs were prepared using mRNA isolated from postnatal day 1 or day 90 rat testis according to the procedure supplied by the manufacturer. Nested PCR procedures were performed to increase the specificity. For GAD67 5′ RACE, the external primer was CCACAGTGCCCTTTGCTTTCCACATC, the internal one was CAAACGCTCCATAAACAGTCGTGCCC. The amplification products of 5′ RACE were cloned into pGEM T-Easy vector and sequenced.
RNA extraction and RT-PCR
Total RNAs from Sprague–Dawley rat testis of different postnatal ages and various tissues were extracted using Trizol, and isolated according to the procedure supplied by the manufacturer. Transcription was performed according to the manufacturer’s instructions. Conditions for PCR amplifications were as follows: initial denaturation at 94°C for 4 min; followed by 30 cycles of 30 s denaturation at 94°C; 30 s annealing at 60°C; and 1 min extension at 72°C. The primer sets used for amplification of different GAD transcripts are illustrated in Fig. 1c. The primer sets used for amplification of different GAD67 variants are illustrated in Fig. 1c. 67A-f, 5′-GCTCCCTGTGGCTGAATCGAG-3′; 67A-r, 5′-CGTCTTGCGGACATAGTTGA-3′; 67B-f, 5′-CCGGGCCGCAGGACCTAGAAG-3′; 67C-f, 5′-GGGAAACTGTAGCCTCTACGC-3′; 67C-r, 5′-GGCCTAGGTGTGTCAACTACTGA-3′; 67-f, 5′-ATTGGTTTAGCTGGCGAATGG-3′; 67-r, 5′-AACAGTCGTGCCTGCGGTTGC-3′. To assure that the results are semi-quantified estimates, β-actin was co-amplified in the system and taken as the internal control.
Determination of transcriptional start sites in rat GAD67 gene. a The PCR products of the 5′ RACE amplification of postnatal day 1 and 90 testis GAD transcripts visualized in 2% agarose gels. Lane Mr DNA marker DL2000. b RT-PCR analysis of GAD67 transcripts in rat testis. Specific primer sets were used, for GAD67A (lane 1) using 67A-f/r, for GAD67B (lane 2) using 67B-f/r, for GAD67C (lane 3) using 67C-f/r, for analyzing alternative splicing of GAD67 transcripts (lane 4) using 67-f/r. Note, two distinct bands were observed when amplified by primer set 67-f/r. c Structural organization of rat GAD67 gene and the transcripts. The exons of GAD67 transcripts are numbered relative to their position in the GAD67 gene (top line). Blue boxes represent the 5′- and 3′-UTR, while the red boxes represent the alternative spliced exon 7. GAD67A was identical to previously reported GAD67 transcript. GAD67B was transcribed from novel TSSs located in intron 2 while GAD67C transcribed from intron 3 located TSSs. GAD67C contains a novel first exon (indicated as ‘T’) located in intron 3 of GAD67 gene. Promoters for GAD67A (Pa), GAD67B (Pb), and GAD67C (Pc) are indicated in the appropriated locations. The ATG codons for translation initiation of each GAD transcript are marked by vertical arrows. Arrows below the exons stand for specific primers used in amplifying the GAD transcripts
Transient transfection and luciferase reporter gene assay
For transient transfection, rat C6 gloma cells and CHO cells were cultured in 24-well tissue culture plates to 80% confluency. The 5′ flanking fragments used to generate the reporter constructs were generated by PCR amplification. The amplified fragments were ligated into the pGL3-basic luciferase reporter vector (Promega). Cells were co-transfected with 1 μg of pGL3-Basic firefly luciferase expression vector containing various length of 5′ flanking sequence of each GAD67 transcripts and 0.075 μg of the internal control pRL-TK vector, which contains Renilla luciferase downstream of the thymidine kinase promoter, using Lipofectamine™ 2000 as described in the manufacturer’s protocol. 48 h after transfection, cells were washed twice with 2 ml of PBS, and cell lysates were prepared with passive lysis buffer in the Dual-Luciferase Reporter Assay System, and firefly and Renilla luciferase activities were measured using a Monolight 2010 luminometer (Analytical Luminescence Laboratory). Luciferase activities are reported as means of values from three independent experiments, each performed at least in duplicate.
Results
Cloning of multiple GAD67 variants with distinct 5′ ends
Rat GAD67 gene is a complex locus consisting of 18 exons and 17 introns, localized to 3q21. To determine the transcriptional start site (TSS) of rat GAD67 gene, 5′ RACE PCR was performed in postnatal day 1 and day 90 rat testes respectively (Fig. 1a). Three transcript variants (GAD67A, GAD67B, and GAD67C) initiated from different TSSs were identified (Fig. 1c). To confirm the expression of GAD67A, GAD67B, and GAD67C in testis, RT-PCR was performed using the primers to anneal the distinct 5′-UTR and ORF: primer set 67A-f and 67A-r for GAD67, primer set 67B-f and 67B-r for GAD67B, and primer set 67C-f and 67C-r for GAD67C. To determine whether alternative exon (exon 7) also present in testis GAD67 transcripts, primer set (67-f and 67-r) that located in common exons 6 and 10 of all three transcripts was designed. This primer may also be used for analyzing total expression level of GAD67 gene. As expected, analysis by agarose gel electrophoresis demonstrated distinct single bands for GAD67A, GAD67B, and GAD67C transcripts. However, two PCR products in size of approximately 453 and 536 bp were observed using primer set (67-f and 67-r) when analyzing mRNA sample of postnatal day 1 testis (Fig. 1b, lane 4). By sequence analysis, the lower band was expectedly confirmed as transcripts without exon 7, while the upper band was identified as splicing variant containing alternative exon 7/B. Thus alternative splicing also exist in postnatal rat testis.
A comparison with the overall structural organization of the rat GAD67 gene demonstrated that GAD67A was almost identical to published rat GAD67 transcript (NM_017007). GAD67B transcribed from novel TSSs that located at the intron 1 of GAD67 gene, about 370 bp upstream of the translation start site (Figs. 1c, 2). The difference between GAD67A and GAD67B was restricted to the 5′-UTR and hence are not expected to affect GAD67 protein structure; however, their potential significance cannot be ignored. The 5′-UTR of eukaryotic mRNA has long been known to play crucial roles in posttranscriptional regulation of gene expression through the modulation of RNA transport [16], translational efficiency [17], and RNA stability [18]. GAD67C (EU791557) transcribed from novel TSSs that located at intron 3 of GAD67 gene. The novel ~212 bp sequence at the 5′-end of the GAD67C transcript derived from a unique first exon (‘exon T’ in Fig. 1c) that located within intron 3 of GAD67 gene. Sequence analysis show both GAD67A and GAD67B translate from the same translate start site and encode 67 kDa full-length GAD67 proteins. Characterization and analysis of GAD67C showed the translation start site of this transcript is located at common exon 5 of GAD67 gene. This novel transcript contained an open reading frame of 1335 nt, which predicted to translate a 50 kDa NH2-terminus truncated GAD67.
Sequence analysis of rat GAD67A, GAD67B, and GAD67C transcripts 5′-flanking regions. The translation start site (ATG) was indicated by asterisks, and “A” is assigned as the nt position of +1. Intronic sequences are in plain lowercase letters, exons sequence in uppercase and framed with gray boxes. The vertical arrows indicate the positions of transcriptional start sites identified by 5′ RACE PCR. The distal transcriptional start sites of each transcript were indicated with right arrow downward. Consensus sequences for putative transcription factor binding sites are shown
Sequence analyzing of 5′-flanking regions of GAD67 transcripts
To aid the identification and characterization of the different promoters, we determined the nucleotide sequence of the 5′-flanking regions of each transcript (Fig. 2), which revealed the presence of putative promoter regions, Pa for GAD67A, Pb for GAD67B, and Pc for GAD67C (Fig. 1c). Sequence analysis shows the 190 bp region preceding the transcriptional start site of GAD67A and the 300 bp region preceding the second exon where TSSs of GAD67B located exhibits high G+C content (about 70%). Putative promoter region of GAD67B also lacks classical TATA box, but contain CCAAT box and multiple Sp1 binding site. For GAD67C, the 5′ flanking sequence also contains a CCAAT box, multiple Sp1 binding sites, and a putative initiator element (Inr).
Differential tissue expression patterns of GAD67 transcripts
Amplified by specific primers, the tissue distribution of GAD67 transcripts was determined by RT-PCR using cDNAs of various rat tissues (Fig. 3). The results showed all transcripts had distinct tissue expression patterns. GAD67A was mainly expressed in brain, lung, ovary, pancreas, testis, and initial segment of the epididymis; Minimal level was also detected in heart, spleen, skeletal muscle, uterus, and corpus epididymis (Fig. 3, GAD67A). Similar expression pattern was observed for GAD67B, but it was not detected in heart and corpus epididymis, and its expression in pancreas was almost undetectable (Fig. 3, GAD67B). For GAD67C, the result showed it was specifically expressed in the testis and not in any other tissues (Fig. 3, GAD67C). Gene expression that has distinct tissue expression pattern is characteristic of existing alternative promoters. The distinct expression pattern of these transcripts indicated GAD67 gene is rigorously regulated by alternative promoters.
Semiquantitative RT-PCR determination of GAD67 transcripts in various rat tissues by using the primer pairs described in Fig. 1c. PCR products for GAD67A, GAD67B, and GAD67C were visualized in 2% agarose gels. The tissues are numbered as follows: 1, brain; 2, heart; 3, kidney; 4, liver; 5, lung; 6, ovary; 7, pancreas; 8, skeletal muscle; 9, spleen; 10, testis; 11, uterus; 12, initial segment of epididymis; 13, corpus epididymis; 14, caudal epididymis. For GAD67A and GAD67B postnatal day 1 testis was used. For GAD67C, postnatal day 90 testis was used. Lane M DNA marker DL2000
Developmental expression of GAD67 transcripts in rat testis
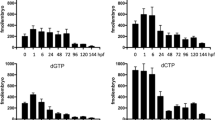
To establish the developmental expression patterns of GAD67 transcripts during postnatal testis maturation, RT-PCR analysis was performed using rat testis cDNA obtained at different time-periods after birth (1–60 days; Fig. 4). When amplified with GAD67A specific primers, the result showed GAD67A was expressed at all age points examined, however, its expression level dramatically decreased after postnatal day 5, then its level gradually increased in the following days (Fig. 4a). When amplified with GAD67B specific primers, we found GAD67B was only expressed in postnatal day 1 and 5, and no expression was detected in the following days (Fig. 4b). Quite different from GAD67A and GAD67B, GAD67C was initially expressed on postnatal day 30 (Fig. 4c). Our results showed all these transcripts have stage specific expression patterns during testis maturation. In order to analyze whether GAD67 gene expression was also regulated by alternative splicing during testis maturation, RT-PCR using primer set (67-f and 67-r) that located in common exons 6 and 10 of all three transcripts was performed. The result showed GAD67 was expressed at all age points examined, but alternative splicing of Exon 7B was only observed in postnatal day 1 and day 5 (Fig. 4d).
Semiquantitative RT-PCR determination of GAD67 transcripts in testis from 1 to 90 postnatal day rats using specific primer pairs described above. β-actin was co-amplified and taken as the internal control. Data are results from three independent experiments and are expressed as mean ± SD. a Developmental expression of GAD67A mRNA. The relative amount of GAD67A transcript was determined by densitometric analysis of amplified DNA band. Expression levels were corrected with β-actin and normalized to the 1st day of age, which was arbitrarily designated as 1. b Developmental expression of GAD67B mRNA. The relative expression level of GAD67B at each postnatal day was analyzed as above. c Developmental expression of GAD67C mRNA. Expression level was normalized to the 60th day of age, which was arbitrarily designated as 1. d Developmental regulated expression of exon 7 and total GAD67 mRNA. Expression level was normalized to the 60th day of age, which was arbitrarily designated as 1
Functional analyzing of 5′-flanking sequences of GAD67 transcripts
To confirm GAD67 transcripts were regulated by independent promoters, we cloned and analyzed the 5′-flanking genome sequences using a luciferase reporter assay. Multiple genome sequences, numbered with respected to the translation start site of GAD67A/B, were amplified by PCR and subcloned into luciferase assay vector pGL3-basic (Fig. 5). The resulting reporter vectors were transfected into rat neuroglial cell line C6 or CHO cell. The C6 cell express endogenous GAD67 gene while CHO cell do not. The transfections showed 5′ flanking region of GAD67A were able to activate transcription of the heterologous reporter gene in both C6 and CHO cell lines (Fig. 5, A1 and A2). When analyzing GAD67B promoter region, luciferase reporter-gene assays were performed using a series of 5′ deletion constructs (Fig. 5, B1–B8). The transfections showed these constructs exhibited significant activity in comparison to the empty pGL3 vectors in C6 cells. However, in CHO cells, 5′-flanking sequences of GAD67B had very low promoter activities, except the region from −2709 to −72 (Fig. 5, B1). When analyzing the promoter region of GAD67C, genomic region from +3989 to +5270 contain 5′-flanking sequence of GAD67C was cloned (Fig. 5C). The transfections showed this genomic region has no significant promoter activity in both C6 and CHO cell (Fig. 5C). The above results confirmed expression of GAD67A and GAD67B are regulated by distinct promoters.
Luciferase promoter-reporter assays. The boundaries of tested constructs for 5′ flanking regions of GAD67A (A1 and A2) and GAD67B (B1–B8) are indicated with respected to the GAD67 genomic sequence (top line). The 5′ flanking region of GAD67C was also cloned (C). The location of the translation start site is described as +1. Numbers on both ends of the lines show the positions of the ends of the genomic fragments. Empty pGL3-basic vector was used as base-line control. Transfection efficiency was determined by cotransfection with the vector, which expresses renilla luciferase under the control of the thymidine kinase promoter. The promoter activity results were normalized to the activity of the internal renilla luciferase control. Constructs were transfected into rat C6 glioma cells or CHO cells, and cellular luciferase activity was measured 48 h later. Data are means ± SE of values from three experiments using the same constructs with assays at least in duplicate
Discussion
GABA is the most important inhibitory neurotransmitter in central nervous system. It was also present in the mammalian testes, where may serve as a paracrine modulator of cell proliferation, migration, and differentiation [1, 19]. Previous studies showed that GABA receptors (GABAA, GABAB, GABAC receptors), and GABA transporter (GAT-1) were exist in rat testis Leydig cells and testicular germ cell [20–24], and testicular GABA appears to be linked to the regulation of steroid synthesis by Leydig cells via GABAA receptors. Moreover, GABA transporter, GAT-1, over expression in testis greatly impaired testis development, which embodied reduced testis mass and slowed spermatogenesis in transgenic mice [25]. Though GABA is important for testis development, the regulation of GABA synthesizing enzyme expression in testis is not well studied. Our previous study showed GAD65 gene contains two TATA-less promoters which are alternative used during rat testis maturation [26]. Here, we further revealed that GAD67 also existed in postnatal testis, and its expression was dynamically regulated by alternative promoters and splicing during postnatal testis maturation.
Transcription initiation is one of the most fundamental cellular processes. The identification of transcriptional start sites leads to the detection of the associated core promoters. As an example, the initiation of transcription of mouse angiotensin-converting enzyme gene in male germ cells starts from an alternate promoter within the 12th intron [27]. Here, three isoforms of GAD67 transcripts (GAD67A, GAD67B, and GAD67C) transcribe from three independent clusters of TSSs were identified. The difference between GAD67A and GAD67B was restricted to the 5′-UTR and hence are not expected to affect GAD67 protein structure; however, their potential significance cannot be ignored. The 5′-UTR of eukaryotic mRNA has long been known to play crucial roles in posttranscriptional regulation of gene expression through the modulation of RNA transport [16], translational efficiency [17], and RNA stability [18]. For GAD67C, the TSSs were mapped in the intron 3, downstream of known translation start site. GAD67C predicted to translate a NH2-terminus truncated GAD with an approximate molecular mass of 50 kDa. Previous research showed the NH2-terminal truncated 44 kDa is enzymatic active [12], thus the predicated 50 kDa GAD may also be functional. However, further study is required to understand the physiological importance of GAD67C in testis.
Gene expression that has distinct tissue and developmental pattern is characteristic of existing alternative promoters [28–30]. Here, we found both GAD67A and GAD67B were co-expressed in brain and many nonneural tissues, except in heart where only GAD67A was expressed (Fig. 3). Quite different from GAD67A and GAD67B, GAD67C was specifically expressed in testis. In addition, all these transcripts have distinct expression pattern during testis maturation (Fig. 4). GAD67A was consistent expressed during postnatal testis development, while GAD67B was only expressed in postnatal day 1 and day 5 testes, and GAD67C was initially expressed on postnatal day 30 when haploid germ cells first appear [31]. The distinct expression pattern of these transcripts indicated GAD67 gene is rigorously regulated by alternative promoters. In addition, we found GAD67 gene expression was also developmental regulated by alternative splicing. Exon 7B which previous reported to be only used in embryonic brain also existed in the testis of postnatal day 1 and day 5. However, alternative splicing was not observed in following days. Exon 7B harbors an in-frame stop codon, resulting in the synthesis of a 25-kDa variant of GAD67, GAD25. GAD25 is enzymatically inactive for lost of binding site for pyridoxal phosphate, the cofactor for GAD. Thus, by alternative splicing, proper GAD activity level may be achieved through generating enzymatically inactive GAD form.
Analysis of genome sequence upstream of TSSs of three GAD67 transcripts revealed presence of putative promoter regions. Sequence analysis shows GAD67A promoter lack the TATA and CCAAT box and has multiple Sp1 binding sites, all characteristics of housekeeping promoters. The importance of the Sp1 binding sites of GAD67A promoter had already been addressed. Mutation of these Sp1 sites could inhibit the GAD67A promoter completely. Our transfections showed GAD67A promoter was not cell type restricted as it was able to activate transcription of the heterologous reporter gene in both C6 and CHO cell lines. Promoter region of GAD67B also lacks the TATA box, but contains CCAAT box and multiple Sp1 binding sites. Promoter activity analysis showed GAD67B promoter was able to activate transcription of the heterologous reporter gene in C6 cell. However, in CHO cell, the promoter activities of the 5′ flanking regions of GAD67B were almost undetectable, except for the construct B1. The promoter activity of construct B1 observed in CHO cell most likely derive from GAD67A promoter since this construct also contain 5′ flanking region of GAD67A. The above results confirmed expression of GAD67A and GAD67B are regulated by distinct promoters.
The 5′ flanking region of GAD67C possesses a putative initiator (Inr) element as well as multiple Sp1 binding sites. The Inr is capable of directing accurate transcription initiation of the Pol II promoter, in combination with the Sp1-binding site or the downstream promoter element in TATA-less promoters [32]. However, the Inr element predicted may be not functional because multiple TSSs were identified, and none of which seemed to be used more frequently than the others (Fig. 2). This predicted binding site should be further confirmed by experiment results, such as electrophoretic mobility shift assay and mutation assay. Transfection experiments showed the 5′ flanking region of GAD67C failed to activate transcription of the heterologous reporter gene both in C6 and CHO cell. This is likely, as GAD67C was only expressed in haploid germ cell of testis, and doesn’t express in both C6 and CHO cells. Thus the promoter activity of GAD67C 5′ flanking sequence may not exhibit in C6 and CHO cells. As lack of cell lines derived form haploid germ cell, the appropriate way to analysis promoter activity of 5′ flanking sequence of GAD67C may be through generating transgenic models.
In conclusion, regulated by alternative promoters and spicing, GAD67 gene has distinct expression pattern during postnatal testis maturation. By activation and regulation of different promoters, distinct transcripts can be generated and their levels regulated independently, and these promoters may be coordinated to achieve the level of GAD67 proper to the maturation of the testis. As GAD is the key enzyme that responsible for GABA synthesizing, thus, by dynamic regulation of GAD gene expression, level of local synthesized GABA can be precisely modulated during testis maturation. The findings reported here will provide a framework for further analysis of GAD67 gene expression, and may aid in designing experiments to unravel the role of GABA in mammalian testis.
References
Frungieri MB, Gonzalez-Calvar SI, Chandrashekar V, Rao JN, Bartke A, Calandra RS (1996) Testicular gamma-aminobutyric acid and circulating androgens in Syrian and Djungarian hamsters during sexual development. Int J Androl 19:164–170
Leroy E, Boyer R, Auburger G et al (1998) The ubiquitin pathway in Parkinson’s disease. Nature 395:451–452
Tillakaratne NJ, Erlander MG, Collard MW, Greif KF, Tobin AJ (1992) Glutamate decarboxylases in nonneural cells of rat testis and oviduct: differential expression of GAD65 and GAD67. J Neurochem 58:618–627
Reetz A, Solimena M, Matteoli M, Folli F, Takei K, De Camilli P (1991) GABA and pancreatic beta-cells: colocalization of glutamic acid decarboxylase (GAD) and GABA with synaptic-like microvesicles suggests their role in GABA storage and secretion. EMBO J 10:1275–1284
Houser CR (1991) GABA neurons in seizure disorders: a review of immunocytochemical studies. Neurochem Res 16:295–308
Seidl R, Cairns N, Singewald N, Kaehler ST, Lubec G (2001) Differences between GABA levels in Alzheimer’s disease and Down syndrome with Alzheimer-like neuropathology. Naunyn Schmiedebergs Arch Pharmacol 363:139–145
Kleppner SR, Tobin AJ (2001) GABA signalling: therapeutic targets for epilepsy, Parkinson’s disease and Huntington’s disease. Expert Opin Ther Targets 5:219–239
Kaufman DL, Erlander MG, Clare-Salzler M, Atkinson MA, Maclaren NK, Tobin AJ (1992) Autoimmunity to two forms of glutamate decarboxylase in insulin-dependent diabetes mellitus. J Clin Invest 89:283–292
Curran-Rauhut MA, Petersen SL (2002) Regulation of glutamic acid decarboxylase 65 and 67 gene expression by ovarian steroids: identification of two functionally distinct populations of GABA neurones in the preoptic area. J Neuroendocrinol 14:310–317
Wei J, Wu JY (2008) Post-translational regulation of l-glutamic acid decarboxylase in the brain. Neurochem Res 33:1459–1465
Bond RW, Wyborski RJ, Gottlieb DI (1990) Developmentally regulated expression of an exon containing a stop codon in the gene for glutamic acid decarboxylase. Proc Natl Acad Sci USA 87:8771–8775
Szabo G, Katarova Z, Greenspan R (1994) Distinct protein forms are produced from alternatively spliced bicistronic glutamic acid decarboxylase mRNAs during development. Mol Cell Biol 14:7535–7545
Ayoubi TA, Van De Ven WJ (1996) Regulation of gene expression by alternative promoters. FASEB J 10:453–460
Landry JR, Mager DL, Wilhelm BT (2003) Complex controls: the role of alternative promoters in mammalian genomes. Trends Genet 19:640–648
Pedersen AA, Videbaek N, Skak K, Petersen HV, Michelsen BK (2001) Characterization of the rat GAD67 gene promoter reveals elements important for basal transcription and glucose responsiveness. DNA Seq 11:485–499
Bi J, Hu X, Loh HH, Wei LN (2003) Mouse kappa-opioid receptor mRNA differential transport in neurons. Mol Pharmacol 64:594–599
Nanbru C, Lafon I, Audigier S et al (1997) Alternative translation of the proto-oncogene c-myc by an internal ribosome entry site. J Biol Chem 272:32061–32066
Chen CY, Del Gatto-Konczak F, Wu Z, Karin M (1998) Stabilization of interleukin-2 mRNA by the c-Jun NH2-terminal kinase pathway. Science 280:1945–1949
Frungieri MB, Gonzalez-Calvar SI, Calandra RS (1996) Influence of photoinhibition on GABA and glutamic acid levels, and on glutamate decarboxylase activity in the testis and epididymis of the golden hamster. Int J Androl 19:171–178
Hu JH, He XB, Wu Q, Yan YC, Koide SS (2002) Subunit composition and function of GABAA receptors of rat spermatozoa. Neurochem Res 27:195–199
Li S, Zhang Y, Liu H, Yan Y, Li Y (2008) Identification and expression of GABAC receptor in rat testis and spermatozoa. Acta Biochim Biophys Sin (Shanghai) 40:761–767
Geigerseder C, Doepner R, Thalhammer A et al (2003) Evidence for a GABAergic system in rodent and human testis: local GABA production and GABA receptors. Neuroendocrinology 77:314–323
He X, Zhang Y, Yan Y, Li Y, Koide SS (2003) Identification of GABABR2 in rat testis and sperm. J Reprod Dev 49:397–402
Hu JH, He XB, Yan YC (2000) Identification of gamma-aminobutyric acid transporter (GAT1) on the rat sperm. Cell Res 10:51–58
Hu JH, Zhang JF, Ma YH et al (2004) Impaired reproduction in transgenic mice overexpressing gamma-aminobutyric acid transporter I (GAT1). Cell Res 14:54–59
Liu H, Li S, Zhang Y, Yan Y, Li Y (2009) Dynamic regulation of glutamic acid decarboxylase 65 gene expression in rat testis. Acta Biochim Biophys Sin (Shanghai) 41:545–553
Howard T, Balogh R, Overbeek P, Bernstein KE (1993) Sperm-specific expression of angiotensin-converting enzyme (ACE) is mediated by a 91-base-pair promoter containing a CRE-like element. Mol Cell Biol 13:18–27
Eddy EM (1998) Regulation of gene expression during spermatogenesis. Semin Cell Dev Biol 9:451–457
Choi E, Lee J, Oh J et al (2007) Integrative characterization of germ cell-specific genes from mouse spermatocyte UniGene library. BMC Genomics 8:256
Davuluri RV, Suzuki Y, Sugano S, Plass C, Huang TH (2008) The functional consequences of alternative promoter use in mammalian genomes. Trends Genet 24:167–177
Malkov M, Fisher Y, Don J (1998) Developmental schedule of the postnatal rat testis determined by flow cytometry. Biol Reprod 59:84–92
Smale ST, Schmidt MC, Berk AJ, Baltimore D (1990) Transcriptional activation by Sp1 as directed through TATA or initiator: specific requirement for mammalian transcription factor IID. Proc Natl Acad Sci USA 87:4509–4513
Acknowledgements
We thank Zhili Wu and Yihong Wang for their excellent technical assistance. This study was supported by grants from the “973” program supported by the Ministry of National Science and Technology, No. 2007CB947100; Shanghai Municipal Commission for Science and Technology, No. 074319111 and No. 07DZ22919.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, H., Zhang, Y., Li, S. et al. Dynamic regulation of glutamate decarboxylase 67 gene expression by alternative promoters and splicing during rat testis maturation. Mol Biol Rep 37, 3111–3119 (2010). https://doi.org/10.1007/s11033-009-9889-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-009-9889-4