Abstract
Cyclooxygenase-2 (COX-2) is an inducible enzyme converting arachidonic acid to prostaglandins and playing important roles in cancer etiology. The −765G>C and 8473T>C polymorphisms have been implicated in cancer risk. However, the results on the association between the two COX-2 polymorphisms and cancer risk are conflicting. To derive a more precise estimation of the association between them, we performed a meta-analysis of 8,090 cancer cases and 11,010 controls concerning −765G>C polymorphism and 14,283 cancer cases and 15,489 controls concerning 8473T>C polymorphism from 33 case–control studies. We used odds ratios (ORs) with 95% confidence intervals (CIs) to assess the strength of the association. Overall, individuals with the −765GC or GC/CC genotypes were associated with higher cancer risk than those with the −765GG genotype and in the stratified analysis this effect maintained in colorectal carcinoma or esophageal cancer of Asian descents. Overall, no significant cancer risk of 8473T>C polymorphism was found. Stratified by cancer types, the variant 8473CC was associated with a decreased risk in breast cancer, compared with the TT or TC/TT genotypes and in lung cancer subgroup after sensitive analysis, there was a decreased risk in CC versus TT, TC versus TT and the dominant models. Moreover, a decreased risk of lung cancer was observed among smokers in the dominant model. In summary, this meta-analysis suggesting that −765G>C may cause an increased risk of colorectal carcinoma and esophageal cancer in Asian descents while 8473T>C polymorphism may cause a decreased risk of breast and lung cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cyclooxygenase-2 (COX-2) is an inducible enzyme that converts arachidonic acid to prostaglandins, which are potent mediators of inflammation. Through the production of prostaglandins, COX-2 is hypothesized to influence carcinogenesis by promoting cell proliferation, inhibiting apoptosis, stimulating angiogenesis, and mediating immune suppression [1–4]. Accumulating evidence has showed that increased expression of COX-2 favors malignant progression [4–7]. There are different polymorphism sites in the COX-2 gene located in 1q25.2–q25.3. Two of these polymorphisms, rs20417(−765G>C) and rs5275(8473T>C) are the most extensively studied polymorphisms, especially in digestive tract cancers and lung cancer respectively.
Recently, there are 19 studies investigated the role of −765G>C polymorphism on the risk of various types of cancer including colorectal carcinoma [8–11], gastric cancer [12, 13], esophageal cancer [14, 15], breast cancer [16, 17], oral cancer [18, 19], prostate cancer [20, 21], and other cancers [22–26] and 23 studies investigated the role of 8473T>C polymorphism on the risk of various types of cancer including lung cancer [27–32], breast cancer [16, 17, 33], prostate cancer [20, 34, 35], esophageal cancer [15, 36], and other cancers [13, 22, 23, 37–39]. However, the results of these studies remain controversial. In consideration of the extensive role of COX-2 in carcinogenesis process, we conduct a meta-analysis on 33 eligible case–control studies to evaluate the association between these two polymorphisms and cancer susceptibility.
Materials and methods
Identification of eligible studies
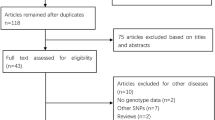
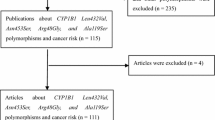
Pubmed and Embase were searched using the search terms (last search was updated 26 March 2009): “PTGS2”, “polymorphism*” and “cancer and/or Neoplasms[Mesh]”. The search was limited to English language papers. All relevant publications were reviewed. And the articles in reference lists were also hand-searched for potentially relevant publications. When more than one of the same or overlapping population was included in several studies, only the most recent or complete study was used for this meta-analysis.
Inclusion criteria
All human-associated studies, regardless of sample size, were included if they met the following criteria: (a) evaluation of −765G>C or 8473T>C polymorphism of COX-2 and cancer risk. (b) case–control studies. (c) sufficient data for examining an Odds ratio (OR) with 95% confidence interval (CI). (d) conforming Hardy–Weinberg equilibrium in the control group.
Data extraction
Two investigators extracted data independently and reached a consensus on all the items. For each study, the following characteristics were collected: the first author’s last name, year of publication, country of origin, ethnicity, matching conditions, numbers of genotyped cases and controls, source of control groups (population- or hospital-based controls), genotyping methods and quality control. Different ethnic descents were categorized as European, Asian, African or Mixed that included more than one ethnic descent. For studies including subjects of different ethnic groups, data were extracted separately for each ethnic group whenever possible [20, 21]. In −765G>C or 8473T>C polymorphism studies, those just presenting the information for genotypes of GG and GC+CC [11, 19] or TT and TC+CC [33], without data for three genotypes, we can only calculate the OR for dominant genetic model. In −765G>C polymorphism studies, overall, there are five studies with zero sample size of CC genotype both in case and control groups, we can only calculate 12 studies under CC versus GG and recessive genetic models. There are two studies [8, 10] with zero sample size of CC genotype both in case and control groups in all three colorectal carcinoma subgroup studies, the effects under CC versus GG and recessive genetic models are not available. There is one study [14] with zero sample size of CC genotype both in case and control groups in all two esophageal cancer subgroup studies, the effects under CC versus GG and recessive genetic models are also not available. There are two studies with zero sample size of CC genotype both in case and control groups concerning oral cancer [18] and pancreatic cancer [26] respectively in all six other cancers subgroup studies, the effects under CC versus GG and recessive genetic models are calculated based on the other four studies. There are five studies with zero sample size of CC genotype both in case and control groups in all eight Asian subgroup studies, the effects under CC versus GG and recessive genetic models are calculated based on the other three studies.
Statistical analysis
The strength of the association between the −765G>C and 8473T>C polymorphisms and cancer risk was measured by ORs with 95% confidence intervals (CIs). The statistical significance of the summary OR was determined with the Z test. For the −765G>C polymorphism, we first estimated the risks of the GC and CC genotypes on cancers, compared with the wild-type GG homozygote. Then risk of CC+GC versus GG and CC versus GC+GG on cancers were evaluated in dominant and recessive effects, respectively. For the 8473T>C polymorphism, we evaluated the same effects. We also carried out the stratified analysis by cancer types and ethnicity (If only one cancer type is included in the meta-analysis, it is combined into the “Other Cancers” group).
Heterogeneity was evaluated by χ2-based Q test among the studies (P < 0.10 was considered significant). When the heterogeneity was present, the random-effects model was used to calculate the pooled ORs [40], whereas the fixed-effects was used in its absence [41]. Sensitivity analyses were performed to assess the stability of the results. Funnel plots was drawn to estimate the potential publication bias, in which the standard error(SE) of log(OR) of each study was plotted against its log(OR). The funnel plot asymmetry was assessed by Egger’s test [42]. The significance of intercept was determined by t-test suggested by Egger, and P < 0.05 was considered representive of statistically significant publication bias.
All statistical test for this meta-analysis were performed with STATA version 9.0 (Stata Corporation College Station, TX), the Review Manager version 4.2 (The Cochrane Collaboration, Oxford, England) and SAS (version 9.1; SAS Institute, Cary, NC).
Results
Study characteristics
A total of 33 studies were retrieved based on the search criteria for cancer susceptibility related to the −765G>C and 8473T>C polymorphisms. The main study characteristics were summarized in Tables 1 and 2. There are 19 case–control studies with 8,090 cancer cases and 11,010 controls concerning −765G>C polymorphism including two studies without the information for all three genotypes either in case or control group [11, 19] and 23 case–control studies with 14,283 cancer cases and 15,489 controls concerning 8473T>C polymorphism including one study [33] just presenting the information for genotypes of TT and TC+CC. For the −765G>C polymorphism, there were 10 studies of Asian descendents with one study [19] just presenting the information for genotypes of GG and GC+CC and one study [11] presenting the information for three genotypes only in control group, seven studies of European descendents without one study [21] because of unconforming of Hardy–Weinberg equilibrium in the control group when separately extracted for the European ethnic group and two studies of African descendents. For the 8473T>C polymorphism, six studies were of Asian descendents, 13 of European descendents and four of mixed descendents with one study [33] just presenting the information for genotypes of TT and TC+CC.
Cancers were confirmed histologically or pathologically in most studies. Of the 33 studies, 17 studies used frequency-matched controls to the cases by the age, sex or ethnicity.
Quantitative synthesis
COX-2 −765G>C
We observed a wide variation of the −765C allele frequencies across different ethnicities. The frequency of −765C allele was 6.01% (95% CI: 1.92–10.11) among Asian controls, which was significantly lower than that in European controls (17.04%; 95% CI: 13.95–20.15, P < 0.000; Table 3, Fig. 1a).
Overall, the variant −765GC heterozygote was associated with a significantly increased risk of all cancer types, compared with the GG (OR = 1.33, 95% CI: 1.10–1.61, P = 0.003, P heterogeneity < 0.00001), and this positive association maintained in colorectal carcinoma (OR = 1.44, 95% CI: 1.09–1.91, P < 0.0001, P heterogeneity = 0.13), esophageal cancer (OR = 2.07, 95% CI: 1.59–2.71, P < 0.00001, P heterogeneity = 0.37), and Asian descents (OR = 1.68, 95% CI: 1.44–1.96, P < 0.00001, P heterogeneity = 0.07) subgroups analyses. Similarly, this significant association maintained under dominant genetic model (CC+GC versus GG) both in overall (OR = 1.32, 95% CI: 1.10–1.58, P = 0.003, P heterogeneity < 0.00001) and colorectal carcinoma (OR = 1.44, 95% CI: 1.09–1.91, P = 0.01, P heterogeneity = 0.14), esophageal cancer (OR = 1.97, 95% CI: 1.51–2.56, P < 0.00001, P heterogeneity = 0.18) and Asian descents (OR = 1.65, 95% CI: 1.42–1.93, P < 0.00001, P heterogeneity = 0.03) subgroups analyses (Table 4). In addition, in colorectal carcinoma and esophageal cancer subgroups, all four and two studies were concerning Asian descents.
Test of heterogeneity
There was significant heterogeneity for heterozygote comparison (GC versus GG: P heterogeneity < 0.00001) and dominant model comparison (CC+GC versus GG: P heterogeneity < 0.00001), but not for homozygote comparison (CC versus GG: P heterogeneity = 0.71) and recessive model comparison (CC versus GC+GG: P heterogeneity = 0.64).
COX-2 8473T>C
We observed a wide variation of the 8473C allele frequencies across different ethnicities. The frequency of 8473C allele was 22.40% (95% CI: 13.46–31.36) among Asian controls, which was significantly lower than that in European controls (34.75%; 95% CI: 32.25–37.26, P < 0.000; Fig. 1b) and Mixed controls (34.72%; 95% CI: 34.20–35.23, P < 0.046; Table 3, Fig. 1b).
We carried out a meta-analysis of COX-2 8473T>C polymorphism in overall, ethnic group and cancer types under various genetic models (Table 5). Stratified by cancer types, the variant 8473CC homozygote was associated with a significantly decreased risk in breast cancer, compared with the TT (OR = 0.84, 95% CI: 0.70–1.00, P = 0.05, P heterogeneity = 0.89) and TC+TT (recessive model) (OR = 0.83, 95% CI: 0.70–0.98, P = 0.03, P heterogeneity = 0.90), the variant 8473TC heterozygote was associated with a significantly decreased risk in other cancers subgroup compared with the TT (OR = 0.86, 95% CI: 0.75–0.98, P = 0.02, P heterogeneity = 0.43). Stratified by ethnicities, the variant 8473CC homozygote was associated with a significantly decreased risk in Mixed descents subgroup, compared with the TT (OR = 0.83, 95% CI: 0.69–0.99, P = 0.04, P heterogeneity = 0.79) and TC+TT (recessive model) (OR = 0.82, 95% CI: 0.69–0.97, P = 0.02, P heterogeneity = 0.84).
Test of heterogeneity
There was significant heterogeneity for homozygote comparison (CC versus TT: P heterogeneity < 0.001), heterozygote comparison (TC versus TT: P heterogeneity = 0.02), dominant model comparison (CC+TC versus TT: P heterogeneity = 0.004) and recessive model comparison (CC versus TC+TT: P heterogeneity < 0.001).
Gene–environment interaction
The data on genotypes of the −765G>C among cases and controls stratified by smoking status were available in three studies that investigated colorectal carcinoma [9], esophageal cancer [14] and pancreatic cancer [26], respectively, while study investigating colorectal carcinoma just presenting the information for genotypes of GG and GC+CC. The smoking status information of esophageal cancer study [14] was extracted from another paper by the same author [43]. There are 928 cases and 1,323 controls in smoker group, and 596 cases and 932 controls in non-smoker group. Among smokers in all three studies, there was a significantly increased cancer risk under dominant model (OR 2.31, 95% CI: 1.63–3.28, P < 0.00001, P heterogeneity = 0.18), but this effect was not present in non-smokers in all three studies. Among both smokers and non-smokers, in esophageal cancer and pancreatic cancer studies, individuals with the GC genotype had a significantly increased cancer risk, compared with the GG genotypes (OR 2.62, 95% CI: 1.79–3.82, P < 0.00001, P heterogeneity = 0.49), (OR 2.10, 95% CI: 1.35–3.26, P = 0.001, P heterogeneity = 0.14), respectively. And this effect was also present under dominant model (Table 4) in these two studies. The results under the CC versus GG and recessive model were unavailable because the sample size of CC in all the two studies was zero in both case and control groups [14, 26].
The data on genotypes of the 8473T>C among cases and controls stratified by smoking status were available in three studies [27, 30, 32] that were all investigating lung cancer, while only one study presenting the information for all three genotypes of TT, TC and CC. So only dominant model could be meta-analyzed, There are 2,379 cases and 1,863 controls in smoker group, and 307 cases and 1,128 controls in non-smoker group and only in smokers in these three studies, there was a significantly decreased cancer risk under dominant model (OR = 0.67, 95% CI: 0.55–0.83, P = 0.0002, P heterogeneity = 0.44) (Table 5).
Sensitivity analyses
COX-2 −765G>C
Sensitivity analyses indicated that only one independent studies by Yang et al. [23] was the main origin of the heterogeneity in the other cancers subgroups. The heterogeneity was effectively removed after exclusion of study by Yang et al. in other cancers subgroup and the effect also had a significant change under both GC versus GG and dominant model (OR = 1.48, 95% CI: 1.07–2.06, P = 0.02, P heterogeneity = 0.01), (OR = 1.45, 95% CI: 1.04–2.02, P = 0.03, P heterogeneity = 0.01), respectively. In addition, no single study influenced the overall OR qualitatively as indicated by sensitivity analyses.
COX-2 8473T>C
Sensitivity analyses indicated that only one independent studies by Campia et al. [30] was the main origin of the heterogeneity in lung cancer subgroup. The heterogeneity was effectively removed after exclusion of study by Campia et al. [30] in lung cancer subgroup and the effect also had a significant change under CC versus TT, TC versus TT and the dominant models (OR = 0.83, 95% CI: 0.70–0.98, P = 0.03, P heterogeneity = 0.55), (OR = 0.88, 95% CI: 0.80–0.97, P = 0.01, P heterogeneity = 0.60) and (OR = 0.87, 95% CI: 0.79–0.96, P = 0.004, P heterogeneity = 0.51), respectively. In addition, no single study influenced the overall OR qualitatively as indicated by sensitivity analyses.
Publication bias
Begger’s funnel plot and Egger’s test were conducted to assess the publication bias of literatures. The shape of funnel plots did not reveal any evidence of funnel plot symmetry. The statistical results still did not show publication bias (P > 0.05, for all).
Discussion
The present meta-analysis 8,090 cases and 11,010 controls concerning the −765G>C polymorphism in the promoter region of COX-2, 14,283 cases and 15,489 controls concerning the 8473T>C polymorphism in the 3′UTR region of COX-2, were included, respectively. And we explored the association between these two potentially functional polymorphisms of COX-2 and cancer risk. We found that the variant heterozygote of the COX-2 −765G>C polymorphism were significantly associated with cancer risk in overall comparisons, compared with the wild homozygote, and the similar significant relationship maintained under dominant genetic model. No significant association between COX-2 8473T>C polymorphism and cancer risk was found under all four genetic models in overall comparisons. COX-2 promoter region contains multiple regulatory elements, such as nuclear factor-κb (NF-κB) binding site, nuclear factor interleukin-6(NF-IL6)/CCAAT/enhancer-binding protein(C/EBP) binding site, cyclic AMP-response element (CRE) and activation protein 1 (AP-1). The regulation of COX-2 gene expression could involve complex interaction among them [44]. As for −765G>C polymorphism of COX-2, conflicting results were reported, previous studies suggested that −765G>C polymorphism in 5′UTR, a potentially functional variant, may eliminate an Sp1-binding site but create an E2F binding site, which results in reduced or increased COX-2 expression [45–47]. There were some studies showed that the 3′UTR of the murine gene for COX-2 contains several regulatory elements altering mRNA stability and translation efficiency [48], which play an important role in degradation, stabilization, and translation of the transcripts [49, 50]. Therefore, polymorphisms in 3′UTR of COX-2 may modify the binding affinity of regulatory factors and alter expression of COX-2, and subsequently influence susceptibility to cancers.
As for −765G>C polymorphism of COX-2, our result showed only in the colorectal carcinoma and esophageal cancer subgroups of Asian descents, under GC versus GG or the dominant model, a significantly high cancer risk was found, while no significant association was found in other cancers under all four genetic models, such as gastric cancer, prostate cancer, breast cancer. Since studies have indicated that COX-2 is up-regulated in various cancers: breast, colon, lung, pancreas, esophagus and prostate [2, 51–56] and the contradictive effects of −765G>C polymorphism on COX-2 expression, the factor that would contribute to this discrepancy is that −765G>C polymorphism might play a different role in different cancers. As for 8473T>C polymorphism of COX-2, in breast cancer subgroup a decreased risk was found under the CC versus TT and the recessive models, in other cancers subgroup under the TC versus TT model a decreased risk was also found, although the association between C allele and lung cancer was inverse, but sensitivity analyses indicated dramatic change when exclusion of study by Campia et al. [29] whose sample size was limited, and it showed that the 8473CC genotype may decrease risk of lung cancer in an C allele dose-response manner. And Since previous study has indicated that COX-2 is up-regulated in breast cancer and lung cancer and a recent study showed that a common SNP (T8473C) in the 3′UTR of the COX-2 gene was shown to be associated with the alteration of mRNA level of the gene, because sequences within the 3′UTR of the COX-2 gene are important for enhancing mRNA translation as well as for translational silencing, we supposed that COX-2 8473T>C polymorphism may reduce cancer risk by translational silencing on post-transcription levels of COX-2, for instance a new target site for MicroRNAs could be created by this polymorphism.
We found an evidence for the association between the −765G>C polymorphism and increased cancer risk among Asians but not among Europeans and Africans under GC versus GG and dominant models. In addition, the 8473T>C polymorphism was associated with an decreased cancer risk among mixed descendents but not among Europeans or Asians in CC versus TT and recessive models, suggesting a possible role of ethnic differences in genetic backgrounds and the environment they lived in [57]. The influence of these two genetic variants may be masked by the presence of other as-yet unidentified causal genes involved in cancer development. Other factors such as selection bias, different matching criteria may also play a role. The above differences may account for the inconsistent results. In addition, there are few reported studies using African populations for COX-2 polymorphisms research. So it is also probably that the observed ethnic differences may due to chance because studies with small sample size may have insufficient statistical power to detect a slight effect. Therefore, additional studies are warranted to further validate ethnic difference in the effect of these two functional polymorphisms on cancer risk, especially in Africans.
Apparent gene–environment interaction was also observed between the −765G>C and tobacco smoking and the risk of cancer was higher among smokers than among non-smokers. While interestingly, a decreased lung cancer risk was found under the dominant model in 8473T>C polymorphism among smokers not among non-smokers (all three studies concerning lung cancer). Studies have shown that COX-2 could be induced by cigarette smoke condensate in vitro and by tobacco-specific-carcinogen-nitrosamine4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone(NNK) in mice [58, 59], which may play an important role in cigarette smoke-induced carcinogenesis. Emerging evidence suggests that COX-2 enzyme plays an important role in lung carcinogenesis [60]. So 8473T>C polymorphism may play a protective effect in lung on reducing high COX-2 levels caused by cigarette smoking among smokers.
Some limitations of this meta-analysis should be addressed. First, lacking of the original data of the reviewed studies limited our further evaluation of potential interactions because the interactions among gene–gene, gene–environment and even different polymorphic loci of the same gene may modulate cancer risk. Second, our result was based on unadjusted estimates, while a more precise analysis should be conducted if more detailed individual data were available, which would allow for an adjusted estimate by other factors such as age and sex. Lacking of the information for the data analysis may cause serious confounding bias. Third, misclassifications on disease status and genotypes may also influence the results because cases in several studies were not confirmed by pathology or other gold standard methods, and the quality control of genotyping was also not well documented in some studies. Fourth, the numbers of published studies were not sufficiently large for a comprehensive analysis, particularly for any given cancer site. In spite of these, our meta-analysis also had some advantages. First, substantial number of cases and controls were pooled from different studies, which significantly increased statistical power of the analysis. Second, the quality of case–control studies included in current meta-analysis was satisfactory and met our inclusion criterion. Third, we did not detect any publication bias indicating that the whole pooled result should be unbiased.
In summary, this meta-analysis provided evidence of the association between the −765G>C and 8473T>C polymorphisms and cancer risk, suggesting that −765G>C may cause an increased risk of colorectal carcinoma and esophageal cancer in Asian descents while 8473T>C polymorphism may cause a decreased risk of breast and lung cancer. However additional large studies are warranted to validate our findings. Future studies should use standardized unbiased genotyping methods and homogeneous cancer patients and well-matched controls and include multi-ethnic groups. Moreover, more sophisticated gene–gene and gene–environment interactions should also be considered in future analysis, which should lead to better, comprehensive understanding of the association between the COX-2 polymorphisms and cancer risk.
Abbreviations
- OR:
-
Odds ratio
- CI:
-
Confidence interval
References
Kirschenbaum A, Liu X, Yao S, Levine AC (2001) The role of cyclooxygenase-2 in prostate cancer. Urology 58:127–131
Fujita H, Koshida K, Keller ET, Takahashi Y, Yoshimito T, Namiki M, Mizokami A (2002) Cyclooxygenase-2 promotes prostate cancer progression. Prostate 53:232–240
Nithipatikom K, Isbell MA, Lindholm PF, Kajdacsy-Balla A, Kaul S, Campell WB (2002) Requirement of cyclooxygenase-2 expression and prostaglandins for human prostate cancer cell invasion. Clin Exp Metastasis 19:593–601
Wang W, Bergh A, Damber JE (2005) Cyclooxygenase-2 expression correlates with local chronic inflammation and tumor neovascularization in human prostate cancer. Clin Cancer Res 11:3250–3256
Fujimura T, Ohta T, Oyama K, Miyashita T, Miwa K (2006) Role of cyclooxygenase-2 in the carcinogenesis of gastrointestinal tract cancers: a review and report of personal experience. World J Gastroenterol 12:1336–1345
van Rees BP, Ristimaki A (2001) Cyclooxygenase-2 in carcinogenesis of the gastrointestinal tract. Scand J Gastroenterol 36:897–903
Marshall SF, Bernstein L, Anton-Culver H, Deapen D, Horn-Ross PL, Mohrenweiser H, Peel D, Pinder R, Purdie DM, Reynolds P et al (2005) Nonsteroidal anti-inflammatory drug use and breast cancer risk by stage and hormone receptor status. J Natl Cancer Inst 97:805–812
Tan W, Wu J, Zhang X, Guo Y, Liu J, Sun T, Zhang B, Zhao D, Yang M, Yu D, Lin D (2007) Associations of functional polymorphisms in cyclooxygenase-2 and platelet 12-lipoxygenase with risk of occurrence and advanced disease status of colorectal cancer. Carcinogenesis 28:1197–1201
Xing LL, Wang ZN, Jiang L, Zhang Y, Xu YY, Li J, Luo Y, Zhang X (2008) Cyclooxygenase-2 polymorphism and colorectal cancer: −765G>C variant modifies risk associated with smoking and body mass index. World J Gastroenterol 14:1785–1789
Hamajima N, Takezaki T, Matsuo K, Saito T, Inoue M, Hirai T, Kato T, Ozeki J, Tajima K (2001) Genotype frequencies of cyclooxygenease 2 (COX2) rare polymorphisms for Japanese with and without colorectal cancer. Asian Pac J Cancer Prev 2:57–62
Koh WP, Yuan JM, van den Berg D, Lee HP, Yu MC (2004) Interaction between cyclooxygenase-2 gene polymorphism and dietary n-6 polyunsaturated fatty acids on colon cancer risk: the Singapore Chinese Health Study. Br J Cancer 90:1760–1764
Pereira C, Sousa H, Ferreira P, Fragoso M, Moreira-Dias L, Lopes C, Medeiros R, Dinis-Ribeiro M (2006) −765G>C COX-2 polymorphism may be a susceptibility marker for gastric adenocarcinoma in patients with atrophy or intestinal metaplasia. World J Gastroenterol 12:5473–5478
Hou L, Grillo P, Zhu ZZ, Lissowska J, Yeager M, Zatonski W, Zhu G, Baccarelli A, Chanock SJ, Fraumeni JF Jr, Chow WH (2007) COX1 and COX2 polymorphisms and gastric cancer risk in a Polish population. Anticancer Res 27:4243–4247
Zhang X, Miao X, Tan W, Ning B, Liu Z, Hong Y, Song W, Guo Y, Shen Y, Qiang B, Kadlubar FF, Lin D (2005) Identification of functional genetic variants in cyclooxygenase-2 and their association with risk of esophageal cancer. Gastroenterology 129:565–576
Upadhyay R, Jain M, Kumar S, Ghoshal UC, Mittal B (2009) Functional polymorphisms of cyclooxygenase-2 (COX-2) gene and risk for esophageal squamous cell carcinoma. Mutat Res 663:52–59
Gao J, Ke Q, Ma HX, Wang Y, Zhou Y, Hu ZB, Zhai XJ, Wang XC, Qing JW, Chen WS, Jin GF, Liu JY, Tan YF, Wang XR, Shen HB (2007) Functional polymorphisms in the cyclooxygenase 2 (COX-2) gene and risk of breast cancer in a Chinese population. J Toxicol Environ Health A 70:908–915
Cox DG, Buring J, Hankinson SE, Hunter DJ (2007) A polymorphism in the 3′ untranslated region of the gene encoding prostaglandin endoperoxide synthase 2 is not associated with an increase in breast cancer risk: a nested case-control study. Breast Cancer Res 9:R3
Chiang SL, Chen PH, Lee CH, Ko AM, Lee KW, Lin YC, Ho PS, Tu HP, Wu DC, Shieh TY, Ko YC (2008) Up-regulation of inflammatory signalings by areca nut extract and role of cyclooxygenase-2-1195G>a polymorphism reveal risk of oral cancer. Cancer Res 68:8489–8498
Lin YC, Huang HI, Wang LH, Tsai CC, Lung O, Dai CY, Yu ML, Ho CK, Chen CH (2008) Polymorphisms of COX-2-765G>C and p53 codon 72 and risks of oral squamous cell carcinoma in a Taiwan population. Oral Oncol 8:798–804
Cheng I, Liu X, Plummer SJ, Krumroy LM, Casey G, Witte JS (2007) COX2 genetic variation, NSAIDs, and advanced prostate cancer risk. Br J Cancer 97:557–561
Panguluri RC, Long LO, Chen W, Wang S, Coulibaly A, Ukoli F, Jackson A, Weinrich S, Ahaghotu C, Isaacs W, Kittles RA (2004) COX-2 gene promoter haplotypes and prostate cancer risk. Carcinogenesis 6:961–966
Vogel U, Christensen J, Wallin H, Friis S, Nexo BA, Tjonneland A (2007) Polymorphisms in COX-2, NSAID use and risk of basal cell carcinoma in a prospective study of Danes. Mutat Res 617:138–146
Yang H, Gu J, Lin X, Grossman HB, Ye Y, Dinney CP, Wu X (2008) Profiling of genetic variations in inflammation pathway genes in relation to bladder cancer predisposition. Clin Cancer Res 14:2236–2244
Peters WH, Lacko M, Te Morsche RH, Voogd AC, Oude Ophuis MB, Manni JJ (2009) COX-2 polymorphisms and the risk for head and neck cancer in white patients. Head Neck 31(7):938–943
Pereira C, Pinto D, Catarino R, Nogal A, Pereira D, Sousa B, Medeiros R (2007) COX-2 polymorphism and susceptibility to gynaecological malignancies: −765C allele confers increased risk for ovarian cancer. Eur J Cancer Suppl 5:314
Zhao D, Xu D, Zhang X, Wang L, Tan W, Guo Y, Yu D, Li H, Zhao P, Lin D (2009) Interaction of cyclooxygenase-2 variants and smoking in pancreatic cancer: a possible role of nucleophosmin. Gastroenterology 136:1659–1668
Hu Z, Miao X, Ma H, Wang X, Tan W, Wei Q, Lin D, Shen H (2005) A common polymorphism in the 3′UTR of cyclooxygenase 2/prostaglandin synthase 2 gene and risk of lung cancer in a Chinese population. Lung Cancer 48:11–17
Park JM, Choi JE, Chae MH, Lee WK, Cha SI, Son JW, Kim CH, Kam S, Kang YM, Jung TH, Park JY (2006) Relationship between cyclooxygenase 8473T<C polymorphism and the risk of lung cancer: a case-control study. BMC Cancer 6:70
Campa D, Zienolddiny S, Maggini V, Skaug V, Haugen A, Canzian F (2004) Association of a common polymorphism in the cyclooxygenase 2 gene with risk of non-small cell lung cancer. Carcinogenesis 25:229–235
Campa D, Hung RJ, Mates D, Zaridze D, Szeszenia-Dabrowska N, Rudnai P, Lissowska J, Fabianova E, Bencko V, Foretova L, Janout V, Boffetta P, Brennan P, Canzian F (2005) Lack of association between polymorphisms in inflammatory genes and lung cancer risk. Cancer Epidemiol Biomarkers Prev 14:538–539
Sorensen M, Autrup H, Tjonneland A, Overvad K, Raaschou-Nielsen O (2005) A genetic polymorphism in prostaglandin synthase 2 (8473, T<C) and the risk of lung cancer. Cancer Lett 226:49–54
Vogel U, Christensen J, Nexø BA, Wallin H, Friis S, Tjonneland A (2007) Peroxisome proliferator-activated receptor2 Pro12Ala, interaction with alcohol intake and NSAID use, in relation to risk of breast cancer in a prospective study of Danes. Carcinogenesis Advance Access originally published online on September 6, 2006 Carcinogenesis 28(2):427–434
Shen J, Gammon MD, Terry MB, Teitelbaum SL, Neugut AI, Santella RM (2006) Genetic polymorphisms in the cyclooxygenase-2 gene, use of nonsteroidal anti-inflammatory drugs, and breast cancer risk. Breast Cancer Res 8:R71
Shahedi K, Lindstrom S, Zheng SL, Wiklund F, Adolfsson J, Sun J, Augustsson-Balter K, Chang BL, Adami HO, Liu W, Gronberg H, Xu J (2006) Genetic variation in the COX-2 gene and the association with prostate cancer risk. Int J Cancer 119:668–672
Danforth KN, Hayes RB, Rodriguez C, Yu K, Sakoda LC, Huang WY, Chen BE, Chen J, Andriole GL, Calle EE, Jacobs EJ, Chu LW, Figueroa JD, Yeager M, Platz EA, Michaud DS, Chanock SJ, Thun MJ, Hsing AW (2008) Polymorphic variants in PTGS2 and prostate cancer risk: results from two large nested case-control studies. Carcinogenesis 29:568–572
Ferguson HR, Wild CP, Anderson LA, Murphy SJ, Johnston BT, Murray LJ, Watson RG, McGuigan J, Reynolds JV, Hardie LJ (2008) Cyclooxygenase-2 and inducible nitric oxide synthase gene polymorphisms and risk of reflux esophagitis, Barrett’s esophagus, and esophageal adenocarcinoma. Cancer Epidemiol Biomarkers Prev 17:727–731
Sakoda LC, Gao YT, Chen BE, Chen J, Rosenberg PS, Rashid A, Deng J, Shen MC, Wang BS, Han TQ, Zhang BH, Cohen-Webb H, Yeager M, Welch R, Chanock S, Fraumeni JF Jr, Hsing AW (2006) Prostaglandin-endoperoxide synthase 2 (PTGS2) gene polymorphisms and risk of biliary tract cancer and gallstones: a population-based study in Shanghai, China. Carcinogenesis 27:1251–1256
Lee TS, Jeon YT, Kim JW, Park NH, Kang SB, Lee HP, Song YS (2007) Lack of association of the cyclooxygenase-2 and inducible nitric oxide synthase gene polymorphism with risk of cervical cancer in Korean population. Ann N Y Acad Sci 1095:134–142
Cox DG, Pontes C, Guino E, Navarro M, Osorio A, Canzian F, Moreno V (2004) Polymorphisms in prostaglandin synthase 2/cyclooxygenase 2 (PTGS2/COX2) and risk of colorectal cancer. Br J Cancer 91:339–343
Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22:719–748
DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7(3):177–188
Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315(7109):629–634
Guo Y, Zhang X, Tan W, Miao X, Sun T, Zhao D, Lin D (2007) Platelet 12-lipoxygenase Arg261Gln polymorphism: functional characterization and association with risk of esophageal squamous cell carcinoma in combination with COX-2 polymorphisms. Pharmacogenet Genomics 17(3):197–205
Wang JM, Ko CY, Chen LC, Wang WL, Chang WC (2006) Functional role of NF-IL6beta and its sumoylation and acetylation modifications in promoter activation of cyclooxygenase 2 gene. Nucleic Acids Res 34:217–231
Papafili A, Hill MR, Brull DJ, McAnulty RJ, Marshall RP, Humphries SE, Laurent GJ (2002) Common promoter variant in cyclooxygenase-2 represses gene expression: evidence of role in acute-phase inflammatory response. Arterioscler Thromb Vasc Biol 22:1631–1636
Brosens LA, Iacobuzio-Donahue CA, Keller JJ, Hustinx SR, Carvalho R, Morsink FH, Hylind LM, Offerhaus GJ, Giardiello FM, Goggins M (2005) Increased cyclooxygenase-2 expression in duodenal compared with colonic tissues in familial adenomatous polyposis and relationship to the −765G → C COX-2 polymorphism. Clin Cancer Res 11:4090–4096
Szczeklik W, Sanak M, Szczeklik A (2004) Functional effects and gender association of COX-2 gene polymorphism G-765C in bronchial asthma. J Allergy Clin Immunol 114:248–253
Cok SJ, Morrison AR (2001) The 3’-untranslated region of murine cyclooxygenase-2 contains multiple regulatory elements that alter message stability and translational efficiency. J Biol Chem 276:23179–23185
Sheng H, Shao J, Dixon DA, Williams CS, Prescott SM, DuBois RN, Beauchamp RD (2000) Transforming growth factor-beta1 enhances Ha-ras-induced expression of cyclooxygenase-2 in intestinal epithelial cells via stabilization of mRNA. J Biol Chem 275:6628–6635
Kuersten S, Goodwin EB (2003) The power of the 3’UTR: translational control and development. Nat Rev Genet 4:626–637
Eberhart CE, Coffey RJ, Radhika A, Giardiello FM, Ferrenbach S, DuBois RN (1994) Up-regulation of cyclooxygenase 2 gene expression in human colorectal adenomas and adenocarcinomas. Gastroenterology 107:1183–1188
Wolff H, Saukkonen K, Anttila S, Karjalainen A, Vainio H, Ristimaki A (1998) Expression of cyclooxygenase-2 in human lung carcinoma. Cancer Res 58:4997–5001
Chan G, Boyle JO, Yang EK et al (1999) Cyclooxygenase-2 expression is up-regulated in squamous cell carcinoma of the head and neck. Cancer Res 59:991–994
Ratnasinghe D, Tangrea J, Roth MJ, Dawsey S, Hu N, Anver M, Wang QH, Taylor PR (1999) Expression of cyclooxygenase-2 in human squamous cell carcinoma of the esophagus; an immunohistochemical survey. Anticancer Res 19:171–174
Hussain T, Gupta S, Mukhtar H (2003) Cyclooxygenase-2 and prostate carcinogenesis. Cancer Lett 191:125–135
Gupta S, Srivastava M, Ahmad N, Bostwick DG, Mukhtar H (2000) Over-expression of cyclooxygenase-2 in human prostate adenocarcinoma. Prostate 42:73–78
Hirschhorn JN et al (2002) A comprehensive review of genetic association studies. Genet 4:45–61
Anto RJ, Mukhopadhyay A, Shishodia S, Gairola CG, Aggarwal BB (2002) Cigarette smoke condensate activates nuclear transcription factor-kappaB through phosphorylation and degradation of IkappaB (alpha), correlation with induction of cyclooxygenase-2. Carcinogenesis 23(9):1511–1518
Hida T, Kozaki K, Ito H, Miyaishi O, Tatematsu Y, Suzuki T et al (2002) Significant growth inhibition of human lung cancer cells both in vitro and in vivo by the combined use of a selective cyclooxygenase 2 inhibitor, JTE-522, and conventional anticancer agents. Clin Cancer Res 8(7):2443–2447
Castelao JE, Bart RD 3rd, DiPerna CA, Sievers EM, Bremner RM (2003) Lung cancer and cyclooxygenase-2. Ann Thorac Surg 76(4):1327–1335
Author information
Authors and Affiliations
Corresponding author
Additional information
Wei Zhu, Bing-bing Wei, Xia Shan and Ping Liu authors have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zhu, W., Wei, Bb., Shan, X. et al. −765G>C and 8473T>C polymorphisms of COX-2 and cancer risk: a meta-analysis based on 33 case–control studies. Mol Biol Rep 37, 277–288 (2010). https://doi.org/10.1007/s11033-009-9685-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-009-9685-1




 ” and “
” and “ ” represent outlier
” represent outlier