Abstract
The pentatricopeptide repeat (PPR) protein family is one of the largest and most complex families in plants. These proteins contain multiple 35-amino acid repeats that are proposed to form a super helix capable of binding RNA. PPR proteins have been implicated in many crucial functions broadly involving organelle biogenesis and plant development. In this study, we identified many genes encoding PPR protein in Upland cotton through an extensive survey of the database of Gossypium hirsutum. Furthermore, we isolated five full-length cDNA of PPR genes from G. hirsutum 0-613-2R which were named GhPPR1–GhPPR5. Domain analysis revealed that the deduced amino acid sequences of GhPPR1–5 contained from 5 to 10 PPR motifs and those PPR proteins were divided into two different PPR subfamilies. GhPPR1–2 belonged to the PLS subfamily and GhPPR3–5 belonged to the P subfamily. Phylogenetic analysis of the five GhPPR proteins and 18 other plant PPR proteins also revealed that the same subfamily clustered together. All five GhPPR genes were differentially but constitutively expressed in roots, stems, leaves, pollens, and fibers based on the gene expression analysis by real-time quantitative RT-PCR. This study is the first report and analysis of genes encoding PPR proteins in cotton.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pentatricopeptide repeat (PPR) proteins comprise a large gene family which was found in a systematic search of the Arabidopsis genome for mitochondrial and chloroplast targeted proteins [1]. PPR proteins are characterized by the signature motif of a degenerate 35-amino acid repeat often arranged in tandem arrays of 2–27 repeats [1, 2]. The PPR protein family is subdivided into two major subfamilies: the P subfamily, whose members contain only canonical PPR motifs, and the PLS subfamily, whose members also contain non-canonical shorter and longer PPR-like motifs [2]. PPR proteins have been found in very small number in animals, fungi, algae; but they are greatly increased in the higher plants. For instance, the Arabidopsis thaliana genome is estimated to have over 440 PPR protein genes and the rice genome encodes over 570 PPR proteins [2, 3]. Evidence from EST databases suggests that many other land plants also contain hundreds of PPR genes [4–6].
The structure of the PPR protein is similar to the related tetratricopeptide (TPR) protein, a 34-amino acid repeating motif which is generally found in eukaryotic proteins [7]. PPR motifs are predicted to consist of a pair of antiparallel α helices, and tandem PPR motifs likely forming a helical binding surface [1]. However, unlike TPR containing proteins which mediate protein–protein interactions [8], PPR proteins are sequence specific RNA-binding proteins. They bind with high affinity to single-stranded RNA, but bind poorly to single or double-stranded DNA [9–12]. Another interesting feature of the PPR proteins is that most of them have an N-terminal sequence predicted to target the protein to either mitochondria or chloroplasts and a number of recent studies reported that PPR proteins play essential roles in regulating the expression of organelle genes. For example, PPR proteins from various higher plants suppress the expression of mitochondrial genes associated with cytoplasmic male sterility [8, 13–15]. They are also associated with transcription [16], translation [17], and many stages of mRNA processing including splicing [18, 19], endonucleolytic cleavage [14] and RNA editing [20, 21]. The growing evidence on PPR protein functions are consistent with the fact that the majority of PPR proteins are predicted to be targeted to mitochondria or chloroplasts.
Cotton is not only the world’s most important natural textile fiber and a significant oilseed crop, but also a crop that is significant for foil energy and bio-energy production. Cotton (Gossypium spp.) belongs to the genus Gossypium of the family Malvaceae. Of the Gossypium species, four are cultivated in agriculture, including two allotetraploids (G. hirsutum and G. barbadense) and two diploids (G. herbaceum and G. arboreum). Gossypium hirsutum, also known as Upland cotton, produces over 95% of the world’s cotton. As with many other plant and animal species of biological and/or economical importance, significant efforts have been made to generate many ESTs and some BAC sequences of cottons. For instance, as of August 25, 2008, 265,833 ESTs were available for the G. hirsutum in GenBank. PPR proteins have been identified in many plants, but up to now there are still no reports on PPR proteins in Upland cotton. Cytoplasmic male sterility (CMS) is a maternally inherited trait which renders plants unable to produce functional pollen; however, male fertility can be restored by nuclear-encoded fertility restorer (Rf) genes. The CMS and Rf systems are a valuable tool in the production of hybrid seed in crop species, including maize, rice and a number of vegetable crops. With the exception of Rf2 from maize, all of the cloned fertility restorer genes so far are members of the PPR gene family [8, 13–15], and the Rf genes of other species are also presumed to encode PPR proteins. In this study, we extensively surveyed the EST and nucleotide databases of G. hirsutum for PPR genes. Moreover, we cloned five PPR genes from Upland cotton 0-613-2R and analyzed their cDNAs encoding PPR proteins. We also investigated the relationship of the five GhPPR genes between the restoring line 0-613-2R and the CMS line 104-7A. The expression patterns of these PPR genes were characterized in different developmental stages by real-time quantitative RT-PCR.
Materials and methods
Plant materials
The Upland cotton lines 0-613-2R and 104-7A used in this study were grown under natural conditions during the growing seasons at the Nanjing Agricultural University.
Preparation of genomic DNA and first strand cDNA synthesis
Genomic DNA was isolated from young cotton leaves using the CTAB method as described previously [22]. Total cellular RNA was extracted from roots, stems and leaves, as well as pollens and fibers using the CTAB-sour phenol extraction method [23]. Each RNA sample was treated with DNase I after the extraction to remove all residual DNA. One microgram of DNA-free total RNA was reverse-transcribed with RevertAid ™ First Stand cDNA Synthesis Kit (MBI) following the manufacturer’s instructions.
Search for genes encoding protein with PPR motifs
Eighteen PPR proteins which have been cloned from Arabidopsis, rice, maize, radish and petunia were used as query sequences for tBLASTn searches of the Gossypium hirsutum EST database (http://www.ncbi.nlm.nih.gov/). Vector sequences and low quality sequences were removed manually from the resulting hits. The remaining non-redundant sequences were assembled by CAP3 assembly tool [24] with default parameter. Potential genes of BAC sequences originated from the G. hirsutum database were predicted by a program: FGENESH (http://www.softberry.com/) and the predicted proteins were used subsequently as input for BLASTP searches against the non-redundant Genbank protein database.
Molecular cloning and sequence analysis of five GhPPR cDNAs
From these gene predictions, gene-specific primers (Table 1) were designed for PCR from cDNA and genomic DNA. The amplification products from both cDNA and genomic DNA were inserted into the pMD18-T vector (TaKaRa, Japan). Sequencing was performed by Nanjing Jinsite Biotechnology Co., Ltd. The PPR domains of the GhPPR proteins were searched with Pfam [25] and InterPro (http://www.ebi.ac.uk/Tools/InterProScan/) [26] and aligned in ClustalX version 1.81 [27]. The export and editing of these sequences were conducted with geneDOC software [28]. The presence of putative mitochondrial localization signals was determined using TargetP [29], Predotar [30], and MitoProt [31].
Semi-quantitative RT-PCR
Semi-quantitative RT-PCR was used to analyze gene expression of 0-613-2R and 104-7A. The gene specific primer pairs (Table 1) were used for PCR reactions under the following conditions: pre-denaturation at 94°C for 5 min, followed by 30 cycles of 30 s at 94°C, 60 s at a specific annealing temperature for each gene (55 or 57°C), and 90 s at 72°C. Cotton EF1a gene, an internal control for constitutive expression, was uniformly expressed in all tissues examined. As an internal control and to exclude genomic contamination, cotton EF1a was amplified (same cycling conditions as above for 28 cycles) from the same cDNA samples.
Real-time quantitative RT-PCR
Real-time quantitative RT-PCR was performed on a Bio-RAD iCycler iQ5Machine. The cotton EF1α gene was amplified as a reference to the target gene for gene expression. PCR products were detected by SYBR Green I fluorescence dye (Invitrogen, USA). A 25 μl PCR amplification mixture contained final concentrations of 1× PCR buffer, 1 mM MgCl2, 0.2 mM dNTPs, 1 μl SYBR Green I (10,000-fold dilution), 1 U rTaq, 0.4 μM of each forward and reverse primers, and cDNA from 25 ng total RNA as template. The following PCR cycling conditions were used: one cycle at 95°C for 3 min, followed by 40 cycles at 94°C for 10 s, 55°C for 20 s and 72°C for 45 s, followed by a final elongation at 72°C for 10 min. Each sample was tested in three replicates.
Phylogenetic analysis of the GhPPR proteins with other plant PPR proteins
Previously published 18 PPR proteins sequences from higher plants were retrieved from the Genbank database together with the five deduced GhPPR proteins sequences for phylogenetic analysis. The phylogenetic tree was constructed with DNAStar software, using MegAlign program.
Results
Searching for ESTs and BACs of G. hirsutum encoding PPR motifs in NCBI database
The allotetraploid G. hirsutum is comprised of two subgenomes (designated AT and DT), and the genome size is estimated to be approximately 2.5 Gb [32]. However, there are only a limited number of genomic sequences available for cotton in GenBank. Up to now, there are 147 BAC sequences of G. hirsutum published on the GenBank totaling about 14.5 Mb. We surveyed these BACs for PPR genes, and found six ORF encoding 2–10 PPR motifs. According to this ratio, there are about 1,034 PPR genes in the allotetraploid G. hirsutum genome with about 517 PPR genes in each subgenome. We also used another method to identify the members of the PPR genes in Upland cotton. Eighteen PPR proteins published from other higher plants were used as queries to screen the G. hirsutum EST database (http://www.ncbi.nlm.nih.gov/). We identified 309 EST contigs encoding PPR motifs from 265,833 G. hirsutum EST database by manually removing the incorrect contigs. Most of the contigs are limited to their 5′ and/or 3′ regions, and therefore the internal sequences are unknown.
Cloning and characterization of the five GhPPR genes
To determine the entire amino acid sequence of the putative PPR proteins, we isolated 5 full-length cDNAs from 3 BAC sequences and 2 EST contigs. Five gene-special primers were designed for RT-PCR from Gossypium hirsutum 0-613-2R, and the products ranged in size from 867 to 1,779 bp (Accession numbers: FJ812358–FJ812367). To identify the intron–exon structure of cotton PPR genes, gene-specific primers were used to amplify the five GhPPR genes from genomic DNA. A comparison between the cDNA and genomic DNA sequences revealed that all the five GhPPR genes contained no intron. This special genomic organization is common to other plant PPR genes.
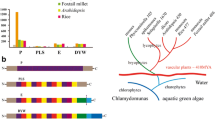
Each of the five GhPPR cDNA sequences contains a single complete ORF encoding protein ranging from 288 to 592 amino acids (Table 2) and these PPR proteins contain 5–10 PPR motifs (Fig. 1a). Further analysis of the PPR domains of the deduced proteins reveals that GhPPR3–5 are typical members of the PPR-P subfamily, and GhPPR1–2 are members of the PLS subfamily which is entirely specific to plants. The GhPPR2 protein also contained a DYW motif in its C-terminal amino acids, which is unique to higher plant PPR proteins and significant for RNA editing [6]. Unlike the E and E+ motifs being highly degenerate, the DYW motifs show a high conservation in the amino acid sequences from the alignment of GhPPR2 and CRR2 (Fig. 1b). Subcellular prediction of the N-terminal amino acids of the five GhPPR proteins show that GhPPR1, GhPPR3 and GhPPR4 are probably targeted to mitochondria and GhPPR2 is probably targeted to chloroplasts, whereas no subcellular signal was detected in GhPPR5 (Table 2).
Schematic diagram of G. hirsutum PPR proteins containing multiple PPR motifs. a PPR motifs are shown as black boxes. Alignments of the PPR motifs of the respective G. hirsutum PPR protein are indicated. Numbers indicate aa positions. The PPR consensus sequences are shaded gray. b Amino acid sequence alignment of the C-terminal DYW motifs of GhPPR2 and CRR2 proteins. Residues identical among the two PPR proteins are shaded black
Expression profiles of GhPPRs in different developmental stages
In order to study the expression pattern of the five GhPPR genes during cotton growth and development, we performed real-time quantitative RT-PCR to measure the relative expression levels in Upland cotton 0-613-2R. The total RNA isolated from a variety of tissues and organs, such as roots, stems, leaves, pollens, and 15dpa fibers was used for further analysis. The results of real-time quantitative RT-PCR showed that each of the five GhPPR genes was expressed ubiquitously in all detected tissues and organs, and this feature was common to the PPR gene family. Although all the five GhPPR genes were expressed constitutively, the relative expression levels were slightly different (Fig. 2). For instance, the transcript accumulation of GhPPR2 was higher in roots and leaves, whereas there was low expression of GhPPR3 in leaves.
Real-time quantitative RT-PCR analysis of five GhPPR transcripts in different organs and tissues in Upland cotton. a–e GhPPR1–GhPPR5, respectively. The samples of various tissues and organs included roots, stems, leaves, pollens and 15dpa fibers in this study. The levels of the five GhPPR mRNA in total RNA samples were analyzed by quantitative real-time RT-PCR using EF1α gene as an internal probe for giving consistently equal levels of samples
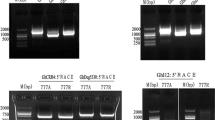
RT-PCR analysis of the five GhPPR genes between the CMS line with its restoring line
With semi-quantitative RT-PCR, we determined whether the five GhPPR genes are potential candidates for the Rf gene of Upland cotton. The specific primers of the five GhPPR genes were used for reverse transcription (RT)-PCR in the pollens of the CMS line 104-7A and the restoring line 0-613-2R (Fig. 4), and the products were recovered for sequencing. However, the cDNA of the five GhPPR genes were identical between the CMS line and the restoring line. Though all cloned Rf genes, except for the Rf2 of maize, encoded PPR proteins, the five GhPPR genes have no relationship with the Rf of cotton, although and they may play other important roles in cotton development.
Phylogenetic analysis of GhPPR proteins and other higher plant PPR proteins
To further characterize the homology and evolutionary relationship of the Upland cotton PPR proteins with other higher plant PPR proteins, we constructed a phylogenetic tree from the amino acid sequences of the five GhPPRs and 18 previously published plant PPR proteins. In this study, overall amino acid sequences were used for phylogenetic analysis, and the phylogenetic tree showed that the GhPPR3, GhPPR4 and GhPPR5 have a close relationship with the members of the P subfamily, while the GhPPR1 and GhPPR2 clustered with members of the PLS subfamily. This result was consistent with the classification of the five GhPPR proteins based on the different PPR domains and C-terminal domains. Interestingly, the Rf genes of different plants clustered into one clade compared with other PPR genes in the P subfamily (Fig. 3).
Discussion
The pentatricopeptide repeat proteins have been identified in almost all eukaryotes, but they are much more abundant in higher plants as compared with other eukaryotes. The Arabidopsis and rice both have several hundreds of PPR genes in their genome, whereas yeast, Drosophila and human genomes are predicted to contain only 5, 2 and 6 PPR genes, respectively [2, 5]. The vast difference between the numbers of PPR genes in higher plants and non-plant organisms indicates that a massive expansion of the PPR gene family occurred during the evolution of plants, but the factors that underlie the expansion of this gene family in plants are not yet understood. In the present study, about 1,034 PPR genes were presumed in the allotetraploid G. hirsutum genome based on six PPR genes in 147 BAC sequences in the Upland cotton database. A predominant feature of the cotton EST set is the significant preference of their tissue sources for fiber or fiber-bearing ovules than other organs. We found 309 PPR unigenes by surveying the G. hirsutum EST database which mostly was from the tissue sources for fiber or fiber-bearing ovules, while only about 24% of them were from non-fiber and non-ovule organs. Therefore, other PPR genes also may be expressed specifically in other tissues and organs. There are as many PPR genes in Upland cotton as in other higher plants, and the increasing number of PPR genes may be correlated with the evolution of land plants towards increasing complexity. Their large number in plants is probably required to meet the special needs of organelle gene expression.
Pentatricopeptide repeat proteins are characterized by tandem arrays of degenerate 35–amino acid motifs [1]. The PPR motif is not one, but in fact three closely related motifs: the canonical PPR motif (P motif), common to all eukaryotes; and two variants specific to plants, the PPR-like S motif (for short) and the PPR-like L motif (for long) [2]. The P subfamily contains only P motifs, whereas the PLS subfamily is comprised of repeats of P-L-S triplets. The plant-specific PPR proteins usually contain additional C-terminal domains E, E+ and DYW motifs in the PLS subfamily and it could be further separated into four subclasses based on their C-terminal domain structure [2]. Whereas E and E+ motifs are highly degenerate and difficult to recognize, DYW motifs contain highly conserved regions including some invariant amino acids which is consistent with our study (Fig. 2b). In our five cloned GhPPR genes, GhPPR3–5 belonged to the classical P subfamily which contained 10, 5 and 7 P motifs, respectively. GhPPR1 and GhPPR2 belonged to the PLS subfamily, with the GhPPR2 was further classified into the DYW subgroup. The DYW domain has not been found in any other protein or in any organism apart from land plants, and it may have catalytic activity for RNA editing due to the presence of invariant cysteine and histidine residues in its motif [2, 6]. The members of this subgroup are predicted to be targeted almost as likely to plastids as to mitochondria. The GhPPR2 is also predicted to target to chloroplast and contains a DYW motif in the C-terminus which suggest GhPPR2 may play an important role in RNA editing in cotton chloroplast. Many functions of the plant PPP proteins are still unknown, but recent studies demonstrated that members of the PPR protein family are involved in the suppression of cytoplasmic male sterility [8, 13–15]. The five GhPPR genes detected by RT-PCR in the CMS line and the restoring carry no differences between them in our study (Fig. 4). Interestingly, the Rf genes from monocot and dicot plants clustered together in the phylogenetic tree (Fig. 3). Therefore, the Rf genes from other plants might be sufficient for future testing of the hypothesis that the monocot and dicot Rf genes share a common ancestor distinct from that of all other PPR genes.
In Arabidopsis, almost half of the plant PPR proteins are predicted to be targeted to mitochondria and one-quarter are predicted to be targeted to chloroplasts [2]. Another peculiarity of PPR genes in Arabidopsis and rice is that the large majority do not contain any introns [2, 5]. We extend these results to Upland cotton in our study. Subcellular prediction of the five deduced GhPPR proteins showed that four of them have subcellular signals in the N-terminus. GhPPR1, GhPPR3 and GhPPR4 are predicted to be targeted to mitochondria and GhPPR2 is probably targeted to chloroplast. From analyses of the genomic organization of the five GhPPR genes, none of them have introns in genome sequences, and this feature may apply to most of the PPR genes in Upland cotton. The majority of PPR genes have been shown to express constitutively in various experiments comparing different plant organs [2, 33]. Similarly, the five GhPPR genes were expressed in all of the tested tissues and organs (Fig. 2). We identified 309 PPR unigenes from the EST database where most of them were sequenced from fiber cDNAs. Although the GhPPR4 and GhPPR5 transcripts originated from those PPR unigenes, they were also expressed in other tissues and organs such as roots, stems and leaves. Therefore, the majority of other GhPPR genes may be expressed constitutively in Upland cotton.
In conclusion, we identified many PPR unigenes in Upland cotton and the characterization and expression pattern of five full-length PPR genes were investigated in our study. Such expression analyses are important for understanding the potential role of these abundant PPR genes in the molecular regulation of the organelle gene expression and plant development. Further studies on these PPR genes in Upland cotton will increase our understanding of the biological significance of the large PPR protein family in land plants.
Abbreviations
- PPR:
-
Pentatricopeptide repeat
- RT-PCR:
-
Reverses transcription polymerase chain
- BAC:
-
Bacterial artificial chromosome
- EST:
-
Expressed sequence tag
- CMS:
-
Cytoplasmic male sterility
References
Small ID, Peeters N (2000) The PPR motif—a TPR-related motif prevalent in plant organellar proteins. Trends Biochem Sci 25:45–47. doi:10.1016/S0968-0004(99)01520-0
Lurin C, Andres C, Aubourg S, Bellaoui M, Bitton F, Bruyere C, Caboche M, Debast C, Gualberto J, Hoffmann B et al (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16:2089–2103. doi:10.1105/tpc.104.022236
André C, Lurin C, Small ID (2007) The multifarious roles of PPR proteins in plant mitochondrial gene expression. Physiol Plant 129:14–22. doi:10.1111/j.1399-3054.2006.00766.x
Hattori M, Hasebe M, Sugita M (2004) Identification and characterization of cDNAs encoding pentatricopeptide repeat proteins in the basal land plant, the moss Physcomitrella patens. Gene 343:305–311. doi:10.1016/j.gene.2004.09.015
O’Toole N, Hattori M, Andres C, Iida K, Lurin C, Schmitz-Linneweber C, Sugita M, Small I (2008) On the expansion of the pentatricopeptide repeat gene family in plants. Mol Biol Evol 25:1120–1128. doi:10.1093/molbev/msn057
Salone V, Rüdinger M, Polsakiewicz M, Hoffmann B, Groth-Malonek M, Szurek B, Small I, Knoop V, Lurin C (2007) A hypothesis on the identification of the editing enzyme in plant organelles. FEBS Lett 581:4132–4138. doi:10.1016/j.febslet.2007.07.075
Blatch GL, Lässle M (1999) The tetratricopeptide repeat: a structural motif mediating protein–protein interactions. Bioessays 21:932–939. doi:10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N
Desloire S, Gherbi H, Laloui W, Marhadour S, Clouet V, Cattolico L, Falentin C, Giancola S, Renard M, Budar F et al (2003) Identification of the fertility restoration locus, Rfo, in radish, as a member of the pentatricopeptide-repeat protein family. EMBO Rep 4:588–594. doi:10.1038/sj.embor.embor848
Tsuchiya N, Fukuda H, Sugimura T, Nagao M, Nakagama H (2002) LRP130, a protein containing nine pentatricopeptide repeat motifs, interacts with a single-stranded cytosine-rich sequence of mouse hypervariable minisatellite Pc-1. Eur J Biochem 269:2927–2933. doi:10.1046/j.1432-1033.2002.02966.x
Williams-Carrier R, Kroeger T, Barkan A (2008) Sequence-specific binding of a chloroplast pentatricopeptide repeat protein to its native group II intron ligand. RNA 14:1930–1941. doi:10.1261/rna.1077708
Meierhoff K, Felder S, Nakamura T, Bechtold N, Schuster G (2003) HCF152, an Arabidopsis RNA binding pentatricopeptide repeat protein involved in the processing of chloroplast psbB-psbT-psbH-petB-petD RNAs. Plant Cell 15:1480–1495. doi:10.1105/tpc.010397
Schmitz-Linneweber C, Williams-Carrier R, Barkan A (2005) RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell 17:2791–2804. doi:10.1105/tpc.105.034454
Bentolila S, Alfonso AA, Hanson MR (2002) A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc Natl Acad Sci USA 99:10887–10892. doi:10.1073/pnas.102301599
Wang Z, Zou Y, Li X, Zhang Q, Chen L, Wu H, Su D, Chen Y, Guo J, Luo D et al (2006) Cytoplasmic male sterility of rice with boro II cytoplasm is caused by a cytotoxic peptide and is restored by two related PPR motif genes via distinct modes of mRNA silencing. Plant Cell 18:676–687. doi:10.1105/tpc.105.038240
Brown GG, Formanova N, Jin H, Wargachuk R, Dendy C, Patil P, Laforest M, Zhang J, Cheung WY, Landry BS (2003) The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. Plant J 35:262–272. doi:10.1046/j.1365-313X.2003.01799.x
Pfalz J, Liere K, Kandlbinder A, Dietz KJ, Oelmuller R (2006) pTAC2, -6, and -12 are components of the transcriptionally active plastid chromosome that are required for plastid gene expression. Plant Cell 18:176–197. doi:10.1105/tpc.105.036392
Pusnik M, Small I, Read LK, Fabbro T, Schneider A (2007) Pentatricopeptide repeat proteins in Trypanosoma brucei function in mitochondrial ribosomes. Mol Cell Biol 27:6876–6888. doi:10.1128/MCB.00708-07
Schmitz-Linneweber C, Williams-Carrier RE, Williams-Voelker PM, Kroeger TS, Vichas A, Barkan A (2006) A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell 18:2650–2663. doi:10.1105/tpc.106.046110
Longevialle AF, Meyer EH, Andres C, Taylor NL, Lurin C, Millar AH, Small ID (2007) The pentatricopeptide repeat gene OTP43 Is required for trans-splicing of the mitochondrial nad1 intron 1 in Arabidopsis thaliana. Plant Cell 19:3256–3265. doi:10.1105/tpc.107.054841
Kotera E, Tasaka M, Shikanai T (2005) A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 433:326–330. doi:10.1038/nature03229
Okuda K, Myouga F, Motohashi R, Shinozaki K, Shikanai T (2007) Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc Natl Acad Sci USA 104:8178–8183. doi:10.1073/pnas.0700865104
Paterson A, Brubaker C, Wendel J (1993) A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol Biol Rep 11:122–127. doi:10.1007/BF02670470
Jiang J, Zhang T (2003) Extraction of total RNA in cotton tissues with CTAB-acidic phenolic method. Cotton Sci 15:166–167
Huang X, Madan A (1999) CAP3: a DNA sequence assembly program. Genome Res 9:868–877. doi:10.1101/gr.9.9.868
Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer ELL et al. (2007) The Pfam protein families database. Nucleic Acids Res gkm960. doi:10.1093/nar/gkm960
Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Biswas M, Bradley P, Bork P, Bucher P et al (2002) InterPro: an integrated documentation resource for protein families, domains and functional sites. Brief Bioinform 3:225–235. doi:10.1093/bib/3.3.225
Thompson J, Gibson T, Plewniak F, Jeanmougin F, Higgins D (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. doi:10.1093/nar/25.24.4876
Nicholas KB, Nicholas HB, Deerfield DW (1997) GeneDoc: analysis and visualization of genetic variation. EMBNEW NEWS 4:14
Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300:1005–1016. doi:10.1006/jmbi.2000.3903
Small I, Peeters N, Legeai F, Lurin C (2004) Predotar: a tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 4:1581–1590. doi:10.1002/pmic.200300776
Manuel G, Claros PV (1996) Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur J Biochem 241:779–786. doi:10.1111/j.1432-1033.1996.00779.x
Hendrix B, Stewart JM (2005) Estimation of the nuclear DNA content of Gossypium species. Ann Bot (Lond) 95:789–797. doi:10.1093/aob/mci078
Oguchi T, Sage-Ono K, Kamada H, Ono M (2004) Genomic structure of a novel Arabidopsis clock-controlled gene, AtC401, which encodes a pentatricopeptide repeat protein. Gene 330:29–37. doi:10.1016/j.gene.2003.11.012
Acknowledgments
This work was financially supported in part by grants from the National Science Foundation in China (30671324), High-tech program 863 (2006AA100105), and the 111 Project (B08025).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yang, L., Zhu, H., Guo, W. et al. Molecular cloning and characterization of five genes encoding pentatricopeptide repeat proteins from Upland cotton (Gossypium hirsutum L.). Mol Biol Rep 37, 801–808 (2010). https://doi.org/10.1007/s11033-009-9610-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-009-9610-7