Abstract
Cinnamate 4-hydroxylase (C4H, EC 1.14.13.11) plays an important role in the phenylpropanoid pathway, which produces many economically important secondary metabolites. A gene coding for C4H, designated as PhC4H (GenBank accession no. DQ211885) was isolated from Parthenocissus henryana. The full-length PhC4H cDNA is 1,747 bp long with a 1,518-bp open reading frame encoding a protein of 505 amino acids, a 40-bp 5′ non-coding region and a 189-bp 3′-untranslated region. Secondary structure of the deduced PhC4H protein consists of 41.78% alpha helix, 15.64% extended strand and 42.57% random coil. The genomic DNA of PhC4H is 2,895 bp long and contains two introns; intron I is 205-bp and intron II is 1,172-bp (GenBank accession no. EU440734). DNA gel blot analysis revealed that there might be a single copy of PhC4H in Parthenocissus henryana genome. By using anchored PCR, a 963-bp promoter sequence was isolated and it contains many responsive elements conserved in the upstream region of PAL, C4H and 4CL including the P-, A-, L- and H-boxes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Phenylpropanoid pathway produces a wide range of secondary metabolites and many of them have beneficial effects on human health. Resveratrol, one of products in the pathway [1], is the basic skeleton of stilbene, which has many biological properties including anti-cancer, antioxidant and anti-aging [2, 3]. Phytochemical investigations showed that the plants in genus of Parthenocissus (Vitaceae family) are rich in stilbene compounds [4, 5]. Though a resveratrol synthase gene has been isolated from Parthenocissus henryana [6], little is known on other genes relative to the biosynthesis of stilbene and needed to be further studied.
Cinnamate 4-hydroxylase (C4H, EC 1.14.13.11), the second enzyme of the resveratrol biosynthesis pathway, catalyzes the hydroxylation of trans-cinnamic acid to para-coumaric acid [7]. Besides, C4H serves to anchor the enzyme complex, formed by the general phenylpropanoid enzymes (including PAL, C4H and 4CL), to the endoplasmic reticulum membrane through the N-terminal hydrophobic region [8].
The first C4H was purified from Jerusalem artichoke [9]. And five C4H genomic DNA sequences have been isolated from hybrid aspen [10], Arabidopsis [11, 12], pea [13] and oilseed rape [14] so far.
To study C4H in Parthenocissus henryana, we successfully isolated a full-length cDNA coding for C4H (PhC4H). To further understand PhC4H gene structure and regulation in Parthenocissus henryana, the genomic DNA sequence and promoter sequence were isolated. DNA gel blot analysis was performed to reveal PhC4H copy number in Parthenocissus henryana genome.
Materials and methods
RNA and DNA isolation
Total RNA was extracted from young leaves of Parthenocissus henryana by Concert Plant RNA Reagent (Invitrogen, USA). Genomic DNA was isolated from young leaves by Plant DNA Extraction Kit (Tiangen, China).
Rapid amplification of 3′ and 5′ cDNA ends of PhC4H
Total RNA isolated from Parthenocissus henryana was reversely transcribed using RP (5′-GCGGTACCCTTTTTTTTTTTTTTTTTT-3′), and followed by PCR amplification with a pair of degenerate primers dC4HFP (5′-ACTGGCT(G/T/C) CAAGT(A/T/G/C)GG(A/T/C)GA(C/T)G-3′) and dC4HRP (5′-CT(A/G)AA(C/T)T G(C/T)CC(A/T)CC(C/T)TTCTC-3′) based on highly conserved sequences in known C4Hs from other species. Amplification conditions were performed as follows: 10 min at 94°C, 5 cycles of 30 s at 94°C, 30 s at 42°C and 1 min at 72°C, and followed by 25 cycles of 30 s at 94°C, 30 s at 55°C and 1 min at 72°C and a final extension of 10 min at 72°C. The PCR products were recovered and cloned into pGEM T Easy Vector (Promega, USA) and sequenced.
According to the sequencing result, gene specific primers for 3′ RACE and 5′ RACE were designed. Total RNA was used as templates to generate the first cDNA strand with RP, and then the 3′cDNA end was amplified by PCR with RP and a gene specific primer C4H-3R (5′-GGAGGATCCGTTGTTCGTGAAGC-3′). For 5′ RACE, total RNA was reversely transcribed using a gene specific primer C4H-5R (5′-GTACTCGAAGCTCTGAGCCAATC-3′), then the 5′ cDNA end was amplified by PCR with 5′ RACE System for Rapid Amplification of cDNA Ends (Gibco-BRL, USA) according to the manual.
Isolation of cDNA and genomic DNA sequences coding for PhC4H by PCR method
A pair of gene specific primers C4HFP-C (5′-GCGGATCCGCCATGGATCTCATACTC-3′) and C4HRP-C (5′-GCTCTAGAGGGAAATTCAAGCTTCATTGG C-3′) was designed based on the sequencing results of 3′ and 5′ cDNA ends. PCR was performed under following conditions: 5 min at 94°C, 30 cycles of 30 s at 94°C, 1 min at 55°C and 2 min at 72°C, and a final extension of 10 min at 72°C. Genomic DNA sequence was amplified by replacing cDNA with genomic DNA.
Bioinformatics analysis
Blast was done on NCBI (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). Molecular weight, isoelectric point and structural analysis of the deduced PhC4H protein were predicated on the ExPASy Proteomics Server (http://www.expasy.ch/). The secondary and tertiary structures were predicated by program GOR IV [15] (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_gor4.html) and program CPHmodels [16] (http://www.cbs.dtu.dk/services/CPHmodels/), respectively. The Swiss-Pdb Viewer (v3.7) program is used for the graphical representation of tertiary protein structure.
DNA gel blot analysis
PhC4H gene copy number was analyzed by DNA gel blot using DIG-labeled probe generated by PCR. Thirty microgram genomic DNA extracted from young leaves was digested with SphI, XbaI (no cut site within the probe) and EcoRI (one recognition site in the probe), respectively. The digested DNA was fractioned on 0.8%-agarose gel electrophoresis and transferred to Hybond-N+ membrane (Amersham Pharmarcia, USA) by capillary transfer. Hybridization and detection were performed with the DIG system under standard conditions described in the manufacturer’s instructions (Roche, Germany).
Isolation of PhC4H promoter sequence by anchored PCR
The experiment was carried out according to a recent study [17]. A linear amplification was performed by PCR in four 40-μl reactions consisting of 4 μl 10 × PCR buffer, 0.5 μl dNTPs (10 mM), 2 μl PC4HRP1 (10 mM) (5′-TGCTCTCCGTACACCGTGAACACC-3′), 2 μg genomic DNA, and 2 units of Ex Taq-polymerase (TaKaRa, Japan). PCR conditions were as follows: 7 min at 94°C, 25 cycles of 30 s at 94°C, 1 min at 62°C and 2 min at 72°C. The PCR products from four reactions were recovered and dissolved in 16.5 μl sterile water for tailing reaction. The tailing mixture contains 5 μl 5 × tailing buffer, 2.5 μl dCTP (2 mM), and 16.5 μl PCR products. After incubation at 94°C for 5 min, the mixture was hold on ice for 2 min and 1 μl terminal deoxynucleotidyl transferase (TaKaRa, Japan) was added to the mixture. Then it was incubated at 37°C for 15 min and finally at 65°C for 10 min. The poly (dC) tailed DNA was amplified using Abridged Anchor Primer (AAP, 5′-GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG-3′) and a gene specific primer PC4HRP2 (5′-CCGGTGAAGATATCGAACACCACG-3′). The PCR products were purified and ligated to pGEM-T Easy Vector (Promega, USA). Transformants were identified by PCR with Abridged Universal Amplification Primer (AUAP, 5′-GGCCACGCGTCGACTAGTAC-3′) and PC4HRP2; the longest fragment was selected for sequencing.
Results
Isolation and analysis of cDNA sequence encoding cinnamate 4-hydroxylase
A cDNA encoding C4H was isolated from Parthenocissus henryana by RT-PCR and RACE approach. First, a pair of degenerate primers based on other known C4Hs was designed and an about 1.2-kb long fragment was amplified from Parthenocissus henryana. According to the sequence, primers for 5′ RACE and 3′ RACE were designed. A 574-bp and an 1,115-bp fragment were obtained by 5′ and 3′ RACE, respectively. The 1,115-bp fragment obtained by 3′ RACE contains part of the open reading fragment and 3′-untranslated region, which contains an 18-bp long poly (A) tail and a putative polyadenylation signal (AATAA) positioned at 28 bp upstream of poly (A) tail.
According to the sequencing results of 5′ and 3′ cDNA ends, gene specific primers were synthesized, and a 1,518-bp long coding region was isolated from Parthenocissus henryana. Blastn result indicated that the coding region of the cDNA shares highest identity (96%) with a Vitis corresponding gene (EMBL accession no. AM468511). Besides, it is identical to many other known C4Hs including C4H from Camptotheca acuminata (AY621152), Citrus paradisi (AF378333), and Gossypium arboreum (AF286647). The result showed that the gene encodes cinnamate 4-hydroxylase, so it was designated as PhC4H.
The PhC4H cDNA encodes a 57.9-kDa protein with an isoelectric point of 9.05. The deduced amino acid sequence contains featured motifs found in plant P450s, including a hydrophobic region near the N-terminus, followed by a proline-rich region (PPGPLPVP) [18], a conserved heme-binding motif PFGVGRRSCPG (conserved motif is PFGXGRRXCXG) at C-terminus [19], and an AAIETT sequence (the threonine-containing binding pocket for the oxygen molecule) [20]. Furthermore, it possesses the C4H conserved residues N302, I371 and K484 that are essential for substrate recognition and orientation [21].
Secondary and tertiary structure of PhC4H protein
The secondary structure of PhC4H protein (Fig. 1), predicated by GOR IV [15], consists of 41.78% alpha helix, 15.64% extended strand and 42.57% random coil. Alpha helices mainly reside at the N-terminus and the middle region of PhC4H protein. Extended strands mainly distribute at the N-terminus and the C-terminus; while random coils distribute at most region of PhC4H protein.
CPHmodels [16] predicated result showed that PhC4H protein (Fig. 2) is a globular protein. Similar to the results reported by Rupasinghe et al. [22], the tertiary structure of PhC4H protein also contains an alpha-domain and a beta-domain.
Analysis of PhC4H DNA sequence
Comparison of PhC4H genomic DNA and cDNA sequences revealed that the genomic DNA contains two introns (Fig. 3), which is consistent with the C4Hs in other plants. Intron I is 205 bp long, interrupted amino acid 262 and Intron II is 1172 bp long, located between amino acid 307and 308. The known C4Hs are different in intron length (Table 1). For example, AtC4H has two small introns (Intron I, 85 bp; Intron II, 220 bp) [12], whereas PsC4H (CYP73A9v1) has two longer introns (Intron I, 162 bp; Intron II, 1726 bp) [13].
The nucleotide sequences of 5′ exon–intron boundaries are AAgt for Intron I and CCgt for Intron II and 3′ sequences are agGA and agGC, which obeys the standard GT-AG rule.
DNA gel blot analysis
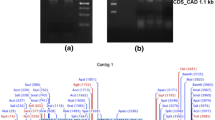
Genomic DNA digested with SphI and XbaI resulted in a single band when hybridized with the probe (Fig. 4). Genomic DNA digested with EcoRI, which has a recognition site within the probe, showed two bands when hybridized with the probe. According to the results, we suggested that PhC4H is encoded by a single locus in Parthenocissus henryana genome.
Analysis of PhC4H promoter
PhC4H promoter is 963 bp long. Nucleotide search showed that it contains the TATA box, which is 70 bp upstream from the translation start codon (ATG), as well as the P-, A- and L-boxes conserved in early phenylpropanoid promoters (Table 2) [23]. PhC4H promoter also contains two Box IV and two G boxes that are involved in light responsiveness, implicating that the expression of PhC4H are regulated by light [24]. In accord with AtC4H and PsC4H, the P- and L-boxes are overlapped in PhC4H promoter [13]. While PhC4H promoter possesses three putative A-boxes, which is absent from AtC4H and PsC4H promoters. The consensus sequence for A-box is CCGTCC, while all the three A-boxes sequence is CCGTCA, with the sixth position C substituted by A in PhC4H promoter.
Discussion
In this work, we have isolated a gene encoding C4H from Parthenocissus henryana. The blastp result demonstrated that the deduced amino acid sequence of PhC4H shares the highest identity with C4H from G. arboreum (up to 89%). Besides, it contains featured sequences found in plant P450 proteins and conserved residues found in C4H proteins.
Previous studies reported that C4H should be grouped into two classes according to different N-terminus and C-terminus [25]. Sequence alignment revealed that PhC4H shows higher identity with the C4Hs of class I than with those of class II (Fig. 5), suggesting that PhC4H should be grouped into class I.
Homology tree of PhC4H and other known C4Hs, showing that PhC4H belongs to class I. C4H from Zinnia elegans (AAB42024), Helianthus tuberosus (CAA78982), Catharanthus roseus (CAA83552), Pisum sativum (AAC49187), Mesembryanthemum crystallinum (AAD11427), Citrus sinensis (AAF66065), Phaseolus vulgaris (CAA70595) and P450s from Persea Americana (CYP71A1), Pleuronectes platessa (CAA52010), Candida maltose (CAA36198), respectively
So far, only five C4H DNA sequences were isolated from hybrid aspen [10], Arabidopsis [11, 12], pea [13] and oilseed rape [14]. All the DNA sequences contain two introns which located at conserved positions. However, they vary in length because of the differences in intron length.
PhC4H is encoded by a single locus, which is consistent with the C4H in Arabidopsis [11, 12] and parsley [26], while C4Hs are encoded by a small gene family in mung bean [27]. C4H plays a key role in both lignin biosynthesis and plant defense in plants which only have one copy of the gene. However, in plants which have two or more copies, each (or some) one plays a specific role in plant physiology and development. For example, three C4Hs in Populus trichocarpa play different physiological roles, PtriC4H1 is involved in the biosynthesis of G lignin, and PtriC4H2 plays a key role in biosynthesis of S lignin, whereas PtriC4H3 participates in stress responses [28].
The secondary structure of PhC4H protein has lower similarity with those of BnC4H proteins [14]. While the tertiary structure of PhC4H protein is similar to those of BnC4H proteins [14], the AtC4H protein and the CYP84A protein (ferulate-5-hydroxylase, a member of plant P450) [22], which accords with the principle that plant P450 proteins possess conserved tertiary structure [29].
It is reported that the genes of general phenylpropanoid pathway are coordinately regulated [13, 23]. PhC4H promoter contains the P-, A-, L-, G- and H-boxes, which were identified in early phenylpropanoid promoters [23], suggesting that the expression of PhC4H might be in accord with PAL and 4CL accumulation. In addition, promoter analysis showed that PhC4H promoter contains MYB-binding sites and a WRKY-binding site. Up to now, several MYB proteins involved in transcriptional regulation of the genes encoding the general phenylpropanoid enzymes have been identified. Previous research showed that AmMYB305 and AmMYB340 positively regulate the expression of PAL [30, 31] and over expression of AmMYB308 and AmMYB330 negatively regulate the expression of C4H in snapdragon (Antirrhinum majus) [32]; while AtMYB4, the first identified MYB protein functions as a transcriptional repressor, plays a negative role in controlling the expression of C4H [33].
Abbreviations
- C4H:
-
Cinnamate 4-hydroxylase
- I:
-
Hypoxanthine
- PAL:
-
Phenylalanine ammonia-lyase
- RACE:
-
Rapid amplification of cDNA ends
- RT-PCR:
-
Reverse transcription polymerase chain reaction
- 4CL:
-
4-Coumarate:coenzyme A ligase
References
Rolfs CH, Kindl H (1984) Stilbene synthase and chalcone synthase: two different constitutive enzymes in cultured cells of Picea excels. Plant Physiol 75:489–492
Jang M, Cai L, Udeani GO, Slowing KV, Thomas CF et al (1997) Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science 275:218–220. doi:10.1126/science.275.5297.218
Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M et al (2004) Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature 430:686–689. doi:10.1038/nature02789
He S, Lu Y, Wu B, Pan Y (2007) Isolation and purification of antioxidative isomeric polyphenols from the roots of Parthenocissus laetevirens by counter-current chromatography. J Chromatogr A 1151:175–179. doi:10.1016/j.chroma.2007.02.102
Son IH, Chung IM, Lee SJ, Moon HI (2007) Antiplasmodial activity of novel stilbene derivatives isolated from Parthenocissus tricuspidata from South Korea. Parasitol Res 101:237–241. doi:10.1007/s00436-006-0454-y
Zhong J, Liu SJ, Ma SS, Yang W, Hu YL, Wu Q et al (2004) Effect of matrix attachment regions on resveratrol production in tobacco with transgene of stilbene synthase from Parthenocissus henryana. Acta Bot Sin 46:948–954
Russell DW (1971) The metabolism of aromatic compounds in higher plants. Properties of cinnamic acid 4-hydroxylase of pea seedlings and some aspects of its metabolic and developmental control. J Biol Chem 246:3870–3878
Winkel-Shirley B (1999) Evidence for enzyme complexes in the phenylpropanoid and flavonoid pathways. Physiol Plant 107:142–149. doi:10.1034/j.1399-3054.1999.100119.x
Gabriac B, Werck-Reichhart D, Teutsch H, Durst F (1991) Purification and immunocharacterization of a plant cytochrome P450: the cinnamic acid 4-hydroxylase. Arch Biochem Biophys 288:302–309. doi:10.1016/0003-9861(91)90199-S
Kawai S, Mori A, Shiokawa T, Kajita S, Katayama Y, Morohoshi N (1996) Isolation and analysis of cinnamic acid 4-hydroxylase homologous genes from a hybrid aspen, Populus kitakamiensis. Biosci Biotechnol Biochem 60:1586–1597
Mizutani M, Ohta D, Sato R (1997) Isolation of a cDNA and a genomic clone encoding cinnamate 4-hydroxylase from Arabidopsis and its expression manner in planta. Plant Physiol 11:755–763. doi:10.1104/pp.113.3.755
Bell-Lelong DA, Cusumano JC, Meyer K, Chapple C (1997) Cinnamate 4-hydroxylase expression in Arabidopsis. Regulation in response to development and the environment. Plant Physiol 113:729–738. doi:10.1104/pp.113.3.729
Whitbred JM, Schuler MA (2000) Molecular characterization of CYP73A9 and CYP82A1 P450 genes involved in plant defense in pea. Plant Physiol 124:47–58. doi:10.1104/pp.124.1.47
Chen AH, Chai YR, Li JN, Chen L (2007) Molecular cloning of two genes encoding cinnamate 4-hydroxylase (C4H) from oilseed rape (Brassica napus). J Biochem Mol Biol 40:247–260
Combet C, Blanchet C, Geourjon C, Deleage G (2000) NIPS@: network protein sequence analysis. Trends Biochem Sci 25:147–150. doi:10.1016/S0968-0004(99)01540-6
Lund O, Nielsen M, Lundegaard C, Worning P (2002) CPHmodels 2.0: X3 M a computer program to extract 3D models. In: Abstract at the CASP5 conference, The Technical University of Denmark, Lyngby, p A102
Chen BJ, Wang Y, Hu YL, Wu Q, Lin ZP (2005) Cloning and characterization of a drought-inducible MYB gene from Boea crassifolia. Plant Sci 168:493–500. doi:10.1016/j.plantsci.2004.09.013
Yamazaki S, Sato K, Suhara K, Sakaguchi M, Mihara K, Omura T (1993) Importance of the proline-rich region following signal anchor sequence in the formation of correct conformation of microsomal cytochrome P450s. J Biochem 114:652–657
Durst F, OKeefe DP (1995) Plant cytochromes P450: an overview. Drug Metabol Drug Interact 12:171–187
Chapple C (1998) Molecular-genetic analysis of plant cytochrome P450-dependent monooxygenases. Annu Rev Plant Physiol Plant Mol Biol 49:311–343. doi:10.1146/annurev.arplant.49.1.311
Schoch GA, Attias R, Le Ret M, Werck-Reichhart D (2003) Key substrate recognition residues in the active site of a plant cytochrome P450, CYP73A1: homology model guided site-directed mutagenesis. FEBS J 270:3684–3695
Rupasinghe S, Baudry J, Schuler MA (2003) Common active site architecture and binding strategy of four phenylpropanoid P450s from Arabidopsis thaliana as revealed by molecular modeling. Protein Eng 16:721–731. doi:10.1093/protein/gzg094
Logemann E, Parniske M, Hahlbrock K (1995) Modes of expression and common structural features of the complete phenylalanine ammonia-lyase gene family in parsley. Proc Natl Acad Sci USA 92:5905–5909. doi:10.1073/pnas.92.13.5905
Menkens AE, Schindler U, Cashmore AR (1995) The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem Sci 20:506–510. doi:10.1016/S0968-0004(00)89118-5
Nedelkina S, Jupe SC, Blee KA, Schalk M, Daniele WR, Bolwell GP (1999) Novel characteristics and regulation of a divergent cinnamate 4-hydroxylase (CYP73A15) from French bean: engineering expression in yeast. Plant Mol Biol 39:1079–1090. doi:10.1023/A:1006156216654
Koopmann E, Logemann E, Hahlbrock K (1999) Regulation and functional expression of cinnamate 4-hydroxylase from parsley. Plant Physiol 119:49–55. doi:10.1104/pp.119.1.49
Mizutani M, Ward E, DiMaio J, Ohta D, Ryals J, Sato R (1993) Molecular cloning and sequencing of a cDNA encoding mung bean cytochrome P450 (P450C4H) possessing cinnamate 4-hydroxylase activity. Biochem Biophys Res Commun 190:875–880. doi:10.1006/bbrc.1993.1130
Lu S, Zhou Y, Li L, Chiang VL (2006) Distinct roles of cinnamate 4-hydroxylase genes in Populus. Plant Cell Physiol 47:905–914. doi:10.1093/pcp/pcj063
Hasemann CA, Kurumbail RG, Boddupalli SS, Peterson JA, Deisenhofer J (1995) Structure and function of cytochromes P450: a comparative analysis of three crystal structures. Structure 3:41–62. doi:10.1016/S0969-2126(01)00134-4
Sablowski RWM, Moyano E, Culianez-Macia FA, Schuch W, Martin C, Bevan M (1994) A flower-specific Myb protein activates transcription of phenylpropanoid biosynthetic genes. EMBO J 13:128–137
Moyano E, Martínez-Garcia JF, Martin C (1996) Apparent redundancy in myb gene function provides gearing for the control of flavonoid biosynthesis in Antirrhinum flowers. Plant Cell 8:1519–1532
Tamagnone L, Merida A, Parr A, Mackay S, Culianez-Macia FA, Roberts K et al (1998) The AmMYB308 and AmMYB330 transcription factors from Antirrhinum regulate phenylpropanoid and lignin biosynthesis in transgenic tobacco. Plant Cell 10:135–154
Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J et al (2000) Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J 19:6150–6161. doi:10.1093/emboj/19.22.6150
Acknowledgments
We are grateful to members of the National Key Laboratory of Protein Engineering and Plant Genetic Engineering at Peking University for comments and discussions. We thank Dr Rongrong Liu for critically reading the manuscript.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, S., Hu, Y., Wang, X. et al. Isolation and characterization of a gene encoding cinnamate 4-hydroxylase from Parthenocissus henryana . Mol Biol Rep 36, 1605–1610 (2009). https://doi.org/10.1007/s11033-008-9357-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-008-9357-6