Abstract
Soybean (Glycine max) is an important oilseed crop that provides ~30% of the vegetable oil used for food, feed and industrial applications. Genetic engineering can produce precise changes in plants in a short period of time and complements conventional plant breeding methods. However, soybean has the lower transformation efficiency and more genotypic limitations than other species, which makes it more difficult to transform. In this study, we introduced diacylglycerolacyl transferase (DGAT) isolated from sesame (Sesamum indicum L.) into the soybean cultivar Dongnong 47, using Agrobacterium-mediated transformation to produce high-oil transgenic lines that are more suitable for breeding. After a 2-year single-location field trial, the transgenic soybean overexpressing SiDGAT1 had increase seed oil content, by an average of over 1.0 percentage point, compared with wild-type plants. Additionally, the transgenic plants expressing SiDGAT1 had significantly reduced protein and soluble sugar contents in mature seeds. SiDGAT1-overexpressing soybean exhibited an altered fatty acid composition, with increases in palmitic (C16:0) and linoleic (C18:2) acid contents and decreases in stearic (C18:0) and oleic (18:1) acid contents, but no major yield change. Thus, engineering the SiDGAT1 enzyme is an effective strategy to improve the oil content and value of soybean.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soybean (Glycine max (L.) Merrill) is an important oilseed crop worldwide that provides 27% of vegetable oil production (Chen et al. 2009). However, the oil content in soybean seeds (20%) is relatively low compared with most other oilseed crops, such as sesame (Sesamum indicum L.; 60%), even though they have a similar lipid biosynthetic pathway (Singh and Hymowitz 1999; Gupta 2008). This suggests that improving the soybean seed oil content through genetic engineering to regulate critical targets and control points in the lipid biosynthetic pathway could be effective (Savadi et al. 2017). Genetic engineering not only can complement conventional plant breeding methods to break yield barriers but also can precisely regulate trait expression in short periods of time without any lineage drag in crop plants (Savadi et al. 2017).
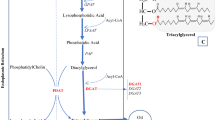
Seed oil is mainly composed of triacylglycerols (TAGs). There are two ways to regulate TAG biosynthesis: through substrate availability, allosteric effectors, and/or enzyme modifications or through the control of lipid-related enzyme synthesis and turnover rates. Diacylglycerolacyl transferase (DGAT) is the only enzyme solely committed to TAG formation, catalyzing diacylglycerol (DAG) from phospholipid synthesis into TAG (Yen et al. 2008; Courchesne et al. 2009). DGAT presents a key rate-limiting enzyme in the control of the flux toward TAG synthesis in the Kennedy pathway (Settlage et al. 1998; Cahoon et al. 2007; Courchesne et al. 2009; Abdullah et al. 2016). Furthermore, the key regulatory role of DGAT in TAG biosynthesis has been established (Guo et al. 2017; Abdullah et al. 2018). The overexpression of the AtDGAT1 gene, driven by the Napin promoter, resulted in an increase of 11–29% in the oil contents of transgenic seeds of Arabidopsis thaliana (Jako et al. 2001). The overexpression of AtDGAT1 and Brassica napus DGAT1 resulted in seed oil content increases of up to 14% in transgenic Arabidopsis compared with the control plants (Weselake et al. 2008). Additionally, field trials of canola, rapeseed (Weselake et al. 2008; Taylor et al. 2009), maize (Zheng et al. 2008; Oakes et al. 2011), and soybean (Lardizabal et al. 2008; Roesler et al. 2016), significant seed oil improvements were achieved by the overexpression of DGAT.
In 2014, we isolated and identified SiDGAT1. The overexpression of SiDGAT1 improved seed oil contents by 1.75 and 1.35 percentage points in T2 and T3 transgenic soybean lines, respectively, in a greenhouse (Wang et al. 2014). However, the receptor soybean cultivar used in that study was Dongnong 50, which has high genetic transformation efficiency, but the seeds are too small for soybean breeding applications. In this study, to breed high seed oil content soybean lines, SiDGAT1 was overexpressed in soybean cultivar Dongnong 47, which is a high oil content cultivar, using the Agrobacterium-mediated cotyledonary node transformation method. Significant increases in seed oil content were achieved by overexpressing SiDGAT1, and a protein content decrease was also observed in two field trials. Moreover, other quality and agronomic traits of transgenic soybean were compared with wild-type soybean.
Materials and methods
Transformation and molecular characterization
To generate SiDGAT1-overexpressing transgenic soybean, the open reading frame of SiDGAT1 was amplified using primer pair 5′-CGTAGATCTTTTAATGGCGATTTTGGAC-3′ and 5′-GTTGGTGACCGGGGTGTAGTATTCATTCCTC-3′ that contained BstEII and BglII restriction enzyme sites, respectively. The amplified product was inserted downstream of the cauliflower mosaic virus (CaMV) 35S promoter of the pCAMBIA3301 vector, replacing the GUS-encoding gene gusA. The recombinant plasmid pCAMBIA3301-SiDGAT1 was introduced into soybean “Dongnong 47” using an Agrobacterium-mediated transformation method described previously (Li et al. 2012a, b; Wang et al. 2014).
T3 generation plants were identified using PCR by selecting bar genes with the primer pair 5′-CCGGCAACAATTAATAGACT-3′ and 5′-TCCATAGTTGCCTGACTCCC-3′ (403 bp). T4 generation plants were identified using PCR tests by selecting bar genes and the 35S promoter with the primer pair 5′-TGTGATAACATGGTGGAGCAC-3′ and 5′-AAATCTCGGTGACGGGCA-3′ (1000 bp).
The T4 generation transgenic plants were further confirmed by Southern blot analysis. Genomic DNA (10 μg) from young leaves of homozygous T4 lines was digested with restriction enzymes EcoRI and HindIII, respectively at 37 °C overnight. Digested DNA was separated on 0.8% (w/v) agarose gels and blotted onto Hybond N+ nylon membrane (Bio-Rad, Hercules, CA, USA) for hybridization with the DIG-labeled bar probe according to the protocol of the DIG-High Prime DNA Labeling and Detection Starter Kit I (Roche, Mannheim, Germany). SiDGAT1 protein expression was tested by western blot analysis in T4 generation plants. The proteins from young leaves were fractionated by dodecyl sulphate-polyarylamide gel electrophoresis (SDS-PAGE) (Laemmli, 1970) using a Mighty Small II electrophoresis system (Hoefer Scientific Instruments, San Francisco, CA, USA). The proteins were resolved on a slab gel (10 × 8 × 0.75 cm), consisting a 13.5% (w/v) separation gel and a 4% (w/v) stacking gel, and electroblotted onto pure nitrocellulose membrane (Midwest-Scientific, Valley Park, MO, USA). Immunoblot analysis was performed using SiDGAT1 protein antibodies made by Abmart Company (Shanghai).
Field trial methods
The T3 and T4 generation transgenic lines “4-1,” “4-3,” “4-4,” and “74-3” that overexpress SiDGAT1 and the “Dongnong 47” soybean (WT) were planted under natural conditions at the Transgenic Experimental Field of Northeast Agricultural University (45° 45′ 06″ N, 126° 43′ 21″ E), Harbin, China in 2017 and 2018. The experiment had a completely randomized block design with three replicates, and plants were planted in rows 3-m long and 0.65-m apart, with 6 cm between plants. Sufficient nutrition and water were supplied throughout the field to avoid potential nutrient and drought stresses. In October, whole seeds were harvested from the plants and air-dried.
In each independent experiment, 10 WT plants and 10 of each transgenic line were randomly sampled at the mature stage. Plant height, number of primary branches, number of primary internode, pod number, seed number per plant, seed weight per plant, and 100-seed weight were measured. Three independent experiments were performed.
The analyses of seed oil and protein contents
Seeds from four transgenic lines and WT were harvested in October. The seed oil content of soybean was analyzed using the Soxhlet extraction method (Tan et al. 2011; Wang et al. 2014). The seed protein content was analyzed using the Kjeldahl method (Leng et al. 2007). The experiments were performed in triplicate, and mean values were used to calculate the oil and protein contents.
Fatty acid composition
Soybean seed powder (0.1 g) was used to analyze the fatty acid composition by gas chromatography (GC-14C, Shimadzu Company, Japan). The fatty acid extraction and analysis were performed according to the method of Zhang et al. (2008). The test columns all had nominal dimensions of 30 m × 0.125 m with a 0.13-μm film thickness. Operating conditions were as follows: carrier, hydric (400 ml/min) split injection, injection temperature 250 °C, detector temperature 250 °C, and column temperature 210 °C (Panthee et al. 2006; Xie et al. 2012). The experiments were performed in triplicate.
Soluble sugar contents
The soluble sugar was measured using anthrone colorimetry (Li et al. 2012a, b). In total, 50 mg soybean powder was used to analyze the soluble sugar contents. The experiments were performed in triplicate.
Statistical analysis
The statistical data analyses were performed using the IBM SPSS Statistics 19 (IBM, Armonk, New York, USA). The significant differences were determined using paired Student’s t tests, and data were presented as means ± standard deviations. Values of p < 0.05 were considered to be statistically significant.
Results
Generation of the transgenic soybean overexpressing SiDGAT1
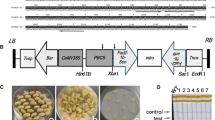
Transgenic soybean plants overexpressing SiDGAT1, driven by the CaMV 35S promoter, were generated by an Agrobacterium-mediated soybean cotyledonary node transformation system. Genomic DNA was isolated from WT and different transgenic plants. The PCR analysis was performed using bar gene-specific primers and Bar+35S promoter-specific primers (Fig. 1a, b). After the PCR analyses of T3 and T4 generation transgenic plants, four independent transgenic lines were obtained. These transgenic lines were further confirmed by Southern blot analysis. The Southern blot data indicated that each of the four transgenic lines contained a single insertion, and a similar single-band pattern was observed in all lines (Fig. 2a). To verify the expression of SiDGAT1 in these transgenic lines, total leaf proteins from the four transgenic lines and WT were separated by SDS-PAGE, and the western blot analysis was performed using antibodies raised against SiDGAT1 (Fig. 2b). The antibody recognized a protein of ~55 kDa from the transgenic soybean leaf, which is similar to the molecular weight of SiDGAT1, while WT produced no reaction. Thus, the data indicated that SiDGAT1 had been introduced into the WT genome, and the SiDGAT1 protein was expressed in transgenic plants. Transgenic lines 4-1, 4-3, 4-4, and 74-3 were chosen for the phenotype analysis.
Identification of transgenic soybean overexpressing SiDGAT1 by PCR analysis. a PCR analysis of T3 generation transgenic lines for the bar gene and 35S promoter. M: DL2000 ladder, 15: positive control (plasmid as template), 14: negative control (H2O as template), 13: negative control (“Dongnong 47” DNA as template), 1–12: transgenic plants. b PCR analysis of T4 generation transgenic lines for the bar gene. M: DL2000 ladder, 11: positive control (plasmid as template), 13: negative control (H2O as template), 12: negative control (“Dongnong 47” DNA as template), 1–10: transgenic plants
Southern and western blot analyses of transgenic soybeans overexpressing SiDGAT1. a Genomic DNA isolated from leaves of wild type and T4 transgenic lines 4-1, 4-2, 4-4, and 74-3 that had been digested with EcoRI or HindIII and separated by agarose gel electrophoresis. M: marker, 1: positive control (plasmid DNA), 2: negative control (“Dongnong 47” DNA), 3–6: DNA from lines 4-1, 4-3, 4-4, and 74-3 digested by EcoRI. 7–12: genomic DNA from lines 4-1, 4-3, 4-4, and 74-3 digested by HindIII. b Total leaf proteins from wild type and T4 transgenic lines (4-1, 4-2, 4-4, and 74-3) fractionated by 13.5% sodium dodecylsulphate-polyarylamide gel electrophoresis and transferred to nitrocellulose membranes. M: markers. 8: negative control. 1–7: transgenic plants
Overexpression of SiDGAT1 significantly increases seed oil content
To determine if the overexpression of SiDGAT1 led to a greater accumulation of seed oil in transgenic soybean, the seed oil contents of four independent transgenic lines were analyzed using the Soxhlet extraction method in 2017 and 2018. An increase in the seed oil content was consistently observed across two generations in a field environment (Fig. 3). In particular, in transgenic lines 4-1 and 4-4, the seed oil content increased by at least 1.0 percentage point compared with WT in the 2017 and 2018 seasons, indicating that this elevated level of seed oil content was retained in the progeny of these transgenic soybean plants. To determine whether SiDGAT1 overexpression would affect fatty acid composition, we compared the fatty acid profiles of the seeds from four transgenic lines with those from WT. As shown in Fig. 4, there were significant increases in palmitic (C16:0) and linoleic (C18:2) acid contents and decreases in stearic (C18:0) and oleic (18:1) acid contents compared with WT. Thus, the overexpression of SiDGAT1 changed the fatty acid composition of transgenic soybean seeds.
Seed oil contents of four SiDGAT1-overexpressing transgenic and wild type plants over two seasons. The data represent the Means ± standard deviations of three replicate experiments, and the values are in dry weight (DW) for seeds. ** indicates significant differences compared with the wild type as determined by Student’s t test (p < 0.01)
Seed fatty acid composition of SiDGAT1-overexpressing transgenic and wild-type plants. The data represent the means ± standard deviations of three replicate experiments, and the values are in dry weight (DW) for seeds. Asterisk (*) indicates significant differences compared with the wild type as determined by Student’s t test (p < 0.05)
Overexpression of SiDGAT1 significantly decreases the seed protein content
With the increase in the seed oil content, a major impact on the protein level was observed in transgenic seeds. SiDGAT1’s overexpression decreased the protein content in transgenic soybean seeds compared with WT (Fig. 5). The decrease in the seed protein content in transgenic lines 4-1, 4-3, and 4-4 were statistically significant (p < 0.01). Thus, the increase in the oil content associated with this transgene produced a correlated decrease in the protein content, in contrast to results obtained from traditional breeding (Burton 1984; Wilcox 1998).
Seed protein contents of four SiDGAT1-overexpressing transgenic and wild-type plants over two seasons. The data represent the means ± standard deviations of three replicate experiments, and the values are in dry weight (DW) for seeds. Double asterisks (**) indicates significant differences compared with the wild type as determined by Student’s t test (p < 0.01)
Overexpression of SiDGAT1 significantly changes seed total soluble sugar content
Mature seeds harvested from the field were subjected to an analysis of the soluble sugar contents. As shown in Fig. 6, the total soluble sugar content decreased significantly (p < 0.01) in the four transgenic lines compared with WT. The greatest reduction occurred in line 4-1, which decreased 6.63 percentage points (43.14%). Therefore, we speculated that the significant improvement in the seed oil content in overexpressing SiDGAT1 soybean was accounted for by the concomitant reductions in the soluble sugar and protein contents.
Seed soluble sugar contents of four SiDGAT1-overexpressing transgenic and wild-type plants. The data represent the means ± standard deviations of three replicate experiments, and the values are in dry weight (DW) for seeds. Double asterisks (**) indicates significant differences compared with the wild type as determined by Student’s t test (p < 0.01)
Field trials
To determine whether the overexpression of SiDGAT1 resulted in growth changes in the transgenic plants, the four SiDGAT1 overexpression lines were grown at the Transgenic Experimental Field of Northeast Agricultural University for 2 years, as was the WT. The data analyses (Table 1) showed that SiDGAT1 overexpression changed the agronomic traits of the transgenic plants. For example, overexpression line 4-1 had a statistically shorter plant height (p < 0.05), but a greater number of primary branches (p < 0.01) and seed number per plant (p < 0.05), as well as a greater seed weight per plant (p < 0.05), compared with WT. The number of primary branches per plant was significantly increased in all transgenic lines compared with WT plants. Seed weight per plant in the transgenic lines had also increased, but it was not statistically significant. Other yield traits did not show statistical changes in comparison with those of WT.
Discussion
DGAT is a very important rate-timing enzyme in the Kennedy pathway. Enhancing DGAT’s activity could improve the carbon flux toward the synthesis of seed-storage lipids (Jako et al. 2001; Taylor et al. 2009). Additionally, the genetic manipulation of DGAT is a promising option for breaking the yield barriers of oilseed crops (Savadi et al. 2017; Wang et al. 2014; Xu et al. 2008; Lardizabal et al. 2008).
Our previous studies have demonstrated that the expression of SiDGAT1 cDNA driven by the CAMV 35S promoter enhanced the seed oil content in transgenic Arabidopsis and soybean grown in a greenhouse (Wang et al. 2014). In this study, we replaced the receptor soybean cultivar Dongnong 50 with Dongnong 47 because the 100-seed weight of the former is just 10.0–11.0 g, which is too small for soybean breeding. The overexpression SiDGAT1 in the Dongnong 47 cultivar also resulted in a significant increase in the transgenic seed oil content. However, the increased oil content in Dongnong 47 is less than that in Dongnong 50. This might be because Dongnong 50 is a low-oil soybean cultivar that has a seed oil content of just 17.54% when grown in the Transgenic Experimental Field of Northeast Agricultural University in 2018. We speculated that the very large increase in the seed oil content of high-oil soybean cultivars might be prevented owing to a “bottleneck,” such as substrate concentration, that likely limits the increase. The overexpression of DGAT1 changes not only the enzyme activity, but also the entire substrate–enzyme–product relationship (Taylor et al. 2009). Additionally, this experiment was conducted in a field, while the previous study was conducted in a greenhouse. The oil content is highly influenced by the environmental conditions (Canvin 1965; Hocking et al. 1997; Fayyaz-ul-Hassan et al. 2005). These two factors may explain why the increase rate in the oil content in Dongnong 47 is less than that in Dongnong 50.
The overexpression of SiDGAT1 in Dongnong 47 increased the palmitic acid (C16:0) and linoleic acid (C18:2) contents and decreased stearic acid (C18:0) and oleic acid (18:1) contents in this study, in marked contrast to that of Roesler et al. (2016). The discrepancies could be due to the DGAT1 from different species. DGAT1 from different species may have the different substrate specificities, and the selectivity of DGAT1 genes varies (Weselake et al. 1991), such as the DGAT1 from B. napus, which prefers 16:0-ACP and very long chain fatty acids, while the DGAT1 of Obroma cacao prefers 18:0-ACP (Giffith and Harwood 1991; Li et al. 2012a, b). DGAT in Arabidopsis has preference for 20:1-CoA, instead of 18:2-CoA and 18:1-CoA (Katavic et al. 1995; Jako et al. 2001). SiDGAT1 comes from sesame, and modified GmDGAT1b comes from soybean. DGAT1 from different species has different enzymatic preferences for substrates during seed development. Hence, DGAT has effects on both oil quantity and quality (Savadi et al. 2016).
With the overexpression of SiDGAT1 leading to a greater than 1.0 percentage point increase in seed oil content, the seed protein and soluble sugar contents suffered significant negative impacts, similar to previous results (Zhang et al. 2005; Savadi et al. 2016; Roesler et al. 2016). Zhang et al. (2005) found that the RNAi-mediated silencing of AtDGAT1 in tobacco led to a reduction in the seed oil content, but increases in protein and sugar contents. Here, the decreases in the protein and soluble sugar contents could have contributed to the increase in the oil contents in transgenic soybean seeds. During seed maturation, seed storage compounds, like starch, oil, and protein, are formed from photosynthates derived from the silique wall and leaves (Meyer and Kinney 2010). Thus, the flow of carbon for TAG synthesis in seed is influence by other metabolic pathways. The overexpression of SiDGAT1 gene in transgenic soybeans increased the assembly of glycerides, and at the same time formed effective competition with other metabolic pathways, such as protein biosynthesis and glucose metabolism, resulting in reduced protein and soluble sugar content in the seeds of transgenic soybeans. And, the reduction in soluble sugars and protein further suggests that seed oil accumulation in SiDGAT1-ox plants is limited by source strength. Additionally, there is a negative correlation between protein and oil contents in soybean (Fillo et al. 2001).
DGAT1’s overexpression did not increase the transgenic soybean yield or 100-seed weight, contrary to previous studies in which the overexpression of DGATs improved seed weights (Zou et al. 1997; Jako et al. 2001; Vigeolas et al. 2007; Wang et al. 2014). In this study, the 100-seed weight of SiDGAT1-overexpression lines did change compared with WT. There was also a small increase in seed weight per transgenic plant, but it was not always statistically significant. Savadi et al. (2016) found no correlation between seed weight and oil content. Thus, breeding for increased yield would not necessarily lead to the overexpression of DGAT, because the high energy content of oil means that a strong positive correlation between yield and oil content is unlikely (Roesler et al. 2016). Additionally, the results from greenhouse experiments are always different compared with those conducted in the natural environment.
The most important finding of this study is that the SiDGAT1-overexpression lines exhibited consistent increases in seed oil contents compared with the WT. Furthermore, the enhanced-seed oil content trend remained consistent. The overexpression of SiDGAT1 can be used to enhance the seed oil content in the Dongnong 47 cultivar, and transgenic soybean plants generated in the study may be used for farming in the future.
References
Abdullah HM, Akbari P, Paulose B, Schnell D, Qi WP, Park Y, Pareek A, Dhankher OP (2016) Transcriptome profiling of Camelina sativa to identify genes involved in triacylglycerol biosynthesis and accumulation in the developing seeds. Biotechnol Biofuels 9:136–155
Abdullah HM, Chhikara S, Akbari P, Schnell DJ, Pareek A, Dhankher OP (2018) Comparative transcriptome and metabolome analysis suggests bottlenecks that limit seed and oil yields in transgenic Camelina sativa expressing diacylglycerol acyltransferase 1 and glycerol-3-phosphate dehydrogenase. Biotechnol Biofuels 11:335–364
Burton JW (1984) Breeding soybeans for improved protein quantity and quality. In: Shibles R (ed) Proceedings of the world soybean research conference III. Westview Press, Boulder, pp 361–367
Cahoon EB, Shockey JM, Dietrich CR, Gidda SK, Mullen RT, Dyer JM (2007) Engineering oilseeds for sustainable production of industrial and nutritional feedstock: solving bottlenecks in fatty acid flux. Curr Opin Plant Biol 10:236–244
Canvin DT (1965) The effect of temperature on the oil content and fatty acid composition of the oils from several oil seed crops. Can J Bot 43(1):63–69
Chen H, Wang FW, Dong YY, Wang N, Sun YP, Li XY, Liu L, Fan XD, Yin HL, Jing YY, Zhang XY, Courchesne NMD, Parisien A, Wang B, Lan CQ (2009) Enhancement of lipid production using biochemical, genetic and transcription factor engineering approaches. J Biotechnol 141:31–41
Courchesne NMD, Parisien A, Wang B, Lan CQ (2009) Enhancement of lipid production using biochemical, genetic and transcription factor engineering approaches. J Biotechnol 141(1–2):31–41
Fayyaz-ul-Hassan AH, Cheema MA, Manaf A (2005) Effects of environmental variation on oil content and fatty acid composition of canola cultivars. J Res (Sci) 16:65–72
Fillo MM, Destro D, Miranda LA, Spinosa WA, Garrao-Panizzi MC, Montalvan R (2001) Relationships among oil content protein content and seed size in soybeans. Braz Arch Biol Technol 44:23–32
Giffith G, Harwood JL (1991) The regulation of triacylglycerol biosynthesis in cocoa (Theobroma cacao L.). Planta 184:27–284
Guo XJ, Fan CM, Chen YH, Wang JQ, Yin WB, Wang RR, Hu ZM (2017) Identification and characterization of an efficient acyl-CoA: diacylglycerol acyltransferase 1 (DGAT1) gene from the miroalga Chlorella ellipsoidea. BMC Plant Biol 17:48–64
Gupta PK (2008) Molecular biology and genetic engineering. Deep and Deep Publications, New Delhi
Hocking PJ, Kirkegaard JA, Angus JF, Gibson AH, Koetz EZ (1997) Comparison of canola Indian mustard and Linola in two contrasting environments: effects of nitrogen fertilizer on dry-matter production, seed yield and seed quality. Field Crop Res 49:107–125
Jako C, Kumar A, Wei Y, Zou J, Barton DL, Giblin EM, Covello PS, Taylor DC (2001) Seed-specific over-expression of an Arabidopsis cDNA encoding a diacylglycerol acyltransferase enhances seed oil content and seed weight. Plant Physiol 126:861–874
Katavic V, Reed DW, Taylor DC, Giblin EM, Barton DL, Zou J, MacKenzie SL, Covello PS, Kunst L (1995) Alteration of seed fatty acid composition by an ethyl methanesulfonate-induced mutation in Arabidopsis thaliana affecting diacylglycerol acyltransferase activity. Plant Physiology 108(1):399–409
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Lardizabal K, Effertz R, Levering C, Mai J, Pedroso MC, Jury T, Aasen E, Gruys K, Bennett K (2008) Expression of Umbelopsis ramanniana DGAT2A in seed increases oil in soybean. Plant Physiol 148:89–96
Leng JT, Chen YZ, Wang Y, Wu CX (2007) Character analysis of newly-developed soybean varieties and breeding objectives in different regions of China. Soybean Sci 26(3):293–299
Li YL, Chen G, Li HY (2012a) Sequence mining and transcript profiling to explore differentially expressed gene associated with lipid biosynthesis during soybean seed development. BMC Plant Biol 12:1
Li R, Yu K, Tateno M, Wu Y, Hatanaka T, Hildebrand D (2012b) Vernonia DGATs can complement the disrupted oil and protein metabolism in epoxygenase-expressing soybean seeds. Metab Eng 14:29–38
Meyer K, Kinney AJ (2010) Biosynthesis and biotechnology of seed lipids including sterols carotenoids and tocochromanols. Lipids Photosynt 30:407–444
Oakes J, Brackenridge D, Colletti R, Daley M, Hawkins DJ, Xiong H, Mai J, Screen SE, Val D, Lardizabal K, Gruys K, Deikman J (2011) Expression of fungal diacylglycerol acyltransferase2 genes to increase kernel oil in maize. Plant Physiol 155(3):1146–1157
Panthee DR, Pantalone VR, Saxton AM (2006) Modifier QTL for fatty acid composition in soybean oil. Euphytica 152:67–73
Roesler K, Shen B, Bermudez E, Li CJ, Hunt J, Damude HG, Ripp KG, Everard JD, Booth JR, Castaneda L, Feng LZ, Meyer K (2016) An improved variant of soybean type 1 diacylglycerol acyltransferase increases the oil content and decreases the soluble carbohydrate content of soybeans. Plant Physiol 171:878–893
Savadi S, Naresh V, Kumar V, Bhat SR (2016) Seed-specific overexpression of Arabidopsis DGAT1 in Indian mustard (Brassica juncea) increases seed oil content and seed weight. Botany 94:177–184
Savadi S, Lambani N, Kashyap PL, Bisht DS (2017) Genetic engineering approaches to enhance oil content in oilseed crops. Plant Growth Regul 83:207–222
Settlage SB, Kwanyuen P, Wilson RF (1998) Relation between diacylglycerol acyltransferase acitivity and oil concentration in soybean. JAOCS 75:775–781
Singh RJ, Hymowitz T (1999) Soybean genetic resources and crop improvement. Genome 42:605–616
Tan HL, Yang XH, Zhang FX, Zheng X, Qu CM, Mu JY, Fu FY, Li JN, Guan RZ, Zhang HS, Wang GD, Oakes J, Brackenridge D, Colletti R, Daley M, Hawkins DJ, Xiong H, Mai J, Screen SE, Val D, Lardizabal K et al (2011) Expression of fungal diacylglycerol acyltransferase2 genes to increase kernel oil in maize. Plant Physiol 155:1146–1157
Taylor DC, Zhang Y, Kumar A, Francis T, Giblin EM, Barton DL, Ferrie JR, Laroche A, Shah S, Zhu W, Snyder CL, Hall L, Rakow G, Harwood JL, Weselake RJ (2009) Molecular modification of triacylglycerol accumulation by over-expression of DGAT1 to produce canola with increased seed oil content under field conditions. Botany 87:533–543
Vigeolas H, Waldeck P, Zank T, Geigenberger P (2007) Increasing seed oil content in oil-seed rape (B. napus L.) by over-expression of a yeast glycerol-3-phosphate dehydrogenase under the control of a seed-specific promoter. Plant Biotechnol J 5:431–441
Wang ZK, Huang WJ, Chang JM, Sebastian A, Li YG, Li HY, Wu XX, Zhang BB, Meng FL, Li WB (2014) Overexpression of SiDGAT1, a gene encoding acyl-CoA: diacylglycerol acyltransferase from Sesamum indicum L. increases oil content in transgenic Arabidopsis and soybean. Plant Cell Tissue Organ Cult 119:399–410
Weselake RJ, Taylor DC, Pomeroy ML, Lawson SL (1991) Properties of diacylglycerol acyltransferase from microspore-derived embryos of Brassica napus L. Phytochemistry 30:3533–3538
Weselake RJ, Shah S, Tang M, Quant PA, Snyder CL, Furukawa-Stoffer TL, Zhu W, Taylor DC, Zou J, Kumar A (2008) Metabolic control analysis is helpful for informed genetic manipulation of oilseed rape (B. napus) to increase seed oil content. J Exp Bot 59:3543–3549
Wilcox JR (1998) Increasing seed protein with eight cycles of recurrent selection. Crop Sci 38:1536–1540
Xie DW, Han YP, Zeng YH, Chang W, Teng WL, Li WL (2012) SSR- and SNP-related QTL underlying linolenic acid and other fatty acid contents in soybean seeds across multiple environments. Mol Breeed 30:169–179
Xu J, Francis T, Mietkiewska E, Giblin EM, Barton DL, Zhang Y, Zhang M, Taylor DC (2008) Cloning and characterization of an acyl-CoA-dependent diacylglycerol acyltransferase 1 (DGAT1) gene from Tropaeolum majus, and a study of the functional motifs of the DGAT protein using site-directed mutagenesis to modify enzyme activity and oil content. Plant Biotechnol J 6(8):799–818
Yen CE, Stone SJ, Koliwad S, Harris C, Farese RV (2008) DGAT enzymes and triacylglycerol biosynthesis. J Lipid Res 49:2283–2301. https://doi.org/10.1194/jlr.R800018-JLR200
Zhang FY, Yang MF, Xu YN (2005) Silencing of DGAT1 in tobacco causes a reduction in seed oil content. Plant Sci 169:689–694
Zhang YJ, Gao HM, Jiang CZ, Hu MY, Liu BQ, Li H (2008) Fast analysis on fatty acids of soybean seed by gas chromatography. Soy Sci 5:859–862
Zheng P, Allen WB, Roseler K, Williams ME, Zhang S, Li J, Glassman K, Ranch J, Nubel D, Solawetz W, Bhattramakki D, Llaca V, Deschamps S, Zhong G, Tarczynski MC, Shen B (2008) A phenylalanine in DGAT is a key determinant of oil content and composition in maize. Nat Genet 40:367–372
Zou JT, Katavic V, Giblin EM, Barton DJ, MacKenzie SL, Keller WA, Hu X, Taylor DC (1997) Modification of seed oil content and acyl composition in the Brassicaceae by expression of a yeast sn-2 acyltransferase gene. Plant Cell 9:909–923
Acknowledgments
We thank Lesley Benyon, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.c/ac), for editing the English text of a draft of this manuscript.
Funding
This study was financially supported by the National Core Soybean Genetic Engineering Project (Contract No. 2016ZX08004003), the “Academic Backbone” Project of Northeast Agricultural University (16XG01), and the Chinese National Natural Science Foundation (30971810).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Wang, Z., Yang, M., Sun, Y. et al. Overexpressing Sesamum indicum L.’s DGAT1 increases the seed oil content of transgenic soybean. Mol Breeding 39, 101 (2019). https://doi.org/10.1007/s11032-019-1016-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-019-1016-1