Abstract
Flowering, an important agronomic trait for seed plants, represents the end of vegetative growth and begins to reproduce. In regulating the floral transition, FLOWERING LOCUS T (FT) encoding a mobile floral signal protein that belongs to the phosphatidylethanolamine-binding protein (PEBP) family acts as critical role. Here, an FT-like gene, DcFT, was isolated and cloned from European Daucus carota L. cultivar “Nantes-H06” (GenBank accession number KY768910), and an alignment of the DcFT protein and other FT-homolog proteins showed that it shared 88.00% similarity with CsFT from Camellia sinensis. Phylogenetic tree analysis indicated that DcFT had the closest relationship with GpFT (Gypsophila paniculata). Quantitative RT-PCR was analyzed to show that the expression pattern of DcFT in inflorescences sharply increased after 10DAA and then slowly increased reaching the maximum at 30DAA. Subcellular localization made clear that the DcFT protein was located in the nucleus and cytoplasm. Broadly speaking, DcFT is an FT-like homologous gene in carrot regulating the floral transition and could be a candidate gene for accelerating the process in carrot breeding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In angiosperms, flowering, the significant agronomic trait in seed plant, is a critical and refined step for plants and is precisely regulated by various endogenous and exogenous (such as environment) factors (Guo et al. 2015; Tan and Swain 2006; Wilkie et al. 2008; Niu et al. 2016). In model plant Arabidopsis thaliana, five primary flowering pathways have been confirmed, incorporating the vernalization, gibberellin, photoperiodic, autonomous, and age pathways (Komeda 2004; Boss et al. 2004; Srikanth and Schimid 2011). FLOWERING LOCUS T (FT) is a vital integrator gene of these flowering pathways and the mobile protein of it encoding induces flowering (Wigge et al. 2005), and the product of the FT gene is widely considered as florigen or the major component of an intricate signal (Zeevaart 2008; Turck et al. 2008; Tamaki et al. 2007; Ahn et al. 2012).
The FT gene was first deciphered in A. thaliana (Kobayashi et al. 1999). Subsequent research revealed that the FT protein was translocated from leaf tissues to the apical meristem and was a direct target of the nuclear protein CONSTANS (CO) in A. thaliana (Samach et al. 2000; Wigge et al. 2005). In long-day (LD) conditions, CO transcription is adjusted by circadian clock, reaching a peak after dawn and activating the downstream TWIN SISTER OF FT (TSF) and FT genes, promoting their expression (Bohlenius et al. 2006). In the apical meristem, the FT protein interacts with FLOWERING LOCUS D (FD), a bZIP transcription factor, forming a complex part that activates the genes LEAFY (LFY), SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1), and APETALA1 (AP1) in the floral meristem to promote flowering (Abe et al. 2005; Ho and Weigel 2014). At present, homologs of FT have been authenticated in many species, including dicotyledonous plants such as sugar beet (Pin et al. 2010), tomato (Lifschitz et al. 2006), soybean (Sun et al. 2011), tobacco (Harig et al. 2012), cucurbits (Lin et al. 2007), onion (Lee et al. 2013), and lettuce (Fukuda et al. 2011), as well as monocots for instance sorghum (Wolabu et al. 2016). This has shown that the FT proteins in various plant species have a conserved functional domain. However, the regulatory mechanism of the vegetative to flowering transition and flower initiation in carrot (Daucus carota L.) has not been reported.
In this study, we cloned the coding region of the “florigen” FT gene from carrot, DcFT, and performed a bioinformatic analysis and expression profile of DcFT in different carrot tissues and organs. To inquire into the subcellular localization of the FT protein in carrot, we fused it to a vector containing green fluorescence protein (GFP) report group and bombarded it into the epidermal cell of onion (Allium cepa L.) by biolistic transformation. Our studies provide a theoretical basis for FT gene in regulating flowering time in carrot (Daucus carota L.) breeding process.
Materials and methods
Plant material and growth condition
The European D. carota cultivared variety “Nantes-H06” was studied in this experiment. Material was cultivated in the experimental field at the Vegetable Research Center of Beijing Agriculture and Forestry Science Academy (Beijing, China). Roots, stems, and leaves were collected at three different periods: before anthesis (no flower primordium), when a distinct meristem (namely, the flower primordium) had formed and when floral organ formation was complete (that is the flower unfolding). Rachises, petioles, and inflorescences (according to the number of days after anthesis (DAA); classified as 10, 20, and 30 DAA) were got from the complete floral organ stage. All materials were harvested and placed in the fridge with − 80 °C for analyzing gene expression levels.
Cloning of DcFT from D. carota
Total RNA was extracted from the leaves of carrot using Trizol reagent (Xinjingke, Beijing, China) according to the slightly modified instruction and then reversed transcription cDNA with a TIANScript RT Kit (Tiangen, Beijing) following the instructions. For cloning the DcFT gene, utilizing the DNAMAN software to contrast with known sequences of the FT homolog from different plants, for instance, Chrysanthemum × morifolium and A. thaliana, designed degenerate primers (FT-M-F and FT-M-R) according to the conservative domains. PCR amplification was done with the product of RT-PCR from the leaves as a template. SMART 5′- and 3′-RACE kits (Clontech, Palo Alto, CA, USA) were used to amplify the DcFT sequence by nested amplification. For 3′-RACE, the antisense primer was from the 3′-RACE kit, and the gene-specific primers were FT-3-out-64 and FT-3-inner-110, respectively. For 5′-RACE, the sense primers were in the 5′-RACE kit and the FT-5-out-269 and FT-5-inner-119 primers were used as inverse primers. The PCR amplification reaction volume and conditions followed the instructions of the SMART 5′- and 3′-RACE kits. FT-GSP-F and FT-GSP-R primers containing restriction enzyme cutting sites were amplified for getting the open reading frame (ORF) that was used for plant expression vector construction. All primers sequences are listed in Supplement Table S1. The products of PCR amplification were checked on 1% denaturing agarose gels and recycled by using a MiniBEST agarose gel DNA extraction kit (Takara, Dalian, China). The PCR product was ligated into the PMD19-T vector (Takara, Dalian, China) and then transformed into Escherichia coli DH5α competent cells (Takara, Dalian, China). After, picking positive clones by using blue-white selection and 5–10 single colonies (white) was sequenced (AuGCT, Beijing, China). Finally, the complete sequences were assembled using DNAMAN to obtain the full-length cDNA of DcFT gene, and then submitted to GenBank.
Bioinformatic and homology comparison of DcFT
The deduced amino acid sequences of carrot DcFT were analyzed using the NCBI blast program (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi). By the online Expasy Protparam tool (http://web.expasy.org/protparam/), the DcFT protein physical properties were predicted. The deduced protein of DcFT and FT proteins from other plant species were aligned using Clustal W (Thompson et al. 1994) with the default parameters. Phylogenetic tree and molecular evolutionary analyses were performed with the MEGA 6.0 software (Neighbor-Joining (NJ) method) (Tamura et al. 2013; Saitou and Nei 1987). Bootstrap evaluation was analyzed with 1000 bootstrap replicates. The numbers represent the bootstrap support (percentage) at each node.
RT-PCR and real-time quantitative PCR
Total RNA (root, stem, leaf, rachis, petiole, and inflorescence (10, 20, and 30 DAA) at the complete floral organ stage and root, stem, and leaf from the other two stages) was extracted by RNAprep pure Plant Kit (Tiangen, Beijing) according to the manufacturer’s protocol. DcFT expression profiles were analyzed by qRT-PCR. The cDNAs were synthesized with the PrimeScript™ RT reagent kit (TaKaRa, Dalian, China) with gDNA Eraser (Perfect Real Time) according to the instructions. The qRT-PCR primers were designed with the Primer Premier 5 software based on the full-length cDNA sequence of the DcFT gene. The β-actin gene (as an internal control) was amplified with the primers to normalize the result (Tian et al. 2015). Before performing the qRT-PCR analysis, the efficiency and specificity of the primers were examined by 1% agarose gel electrophoresis and a preliminary qRT-PCR experiment. The reaction for each sample was repeated in triplicate. The reaction volume was 20 μL, including 2.0 μL cDNA template, 0.4 μL of each primer (10 μM), 7.2 μL PCR-grade water, and 10 μL of SYBR premix Ex Taq II (Perfect Real Time; TaKaRa, Dalian, China). qRT-PCR was performed on a Roche Light Cycler 480 system (Bio-Rad, USA). The program for qRT-PCR was 95 °C for 15 min, followed by 40 cycles of 95 °C for 10 s, 60 °C for 20 s, and 72 °C for 20 s. To confirm the credibility of the qRT-PCR results, a melting curve analysis was performed at the end of each PCR reaction at 95 °C for 5 s, 65 °C for 60 s, and 97 °C continuously, prior to termination at 40 °C for 30 s. The dates were analyzed according to the 2−ΔΔCt methods (Schmittgen and Livak 2008).
Construction of the expression vector
Using a transient expression vector pCambia1305-35S-GFP supplied by Jian Ma (Vegetable Research Center, Beijing) analyzed the transient expression of DcFT gene. To construct the expression vector 35S::DcFT-GFP, the primers containing XbaI and BamHI sites were designed to amplify the DcFT ORF domain. The PCR production was separated on 1% agarose gel and then purified. The empty vector pCambia1305-35S-GFP was digested with restriction enzyme XbaI and BamHI at 37 °C for 3 h. Subsequently, the purified PCR product and cleaved 35S::GFP vector were fused using an In-Fusion HD Cloning kit (TaKaRa, Dalian, China) and then transformed into E. coli DH5α competent cells. Positive recombinants were selected and confirmed by sequencing. Ultimately, the 35S::DcFT-GFP vector was obtained.
Subcellular localization analysis
The obtained 35S::DcFT-GFP fusion proteins were surveyed by biolistic bombardment of onion epidermal cell as the previous method (Scott et al. 1999). The constructed 35S::DcFT-GFP DNA was extracted with an EndoFree Maxi Plasmid Kit (Tiangen, Beijing). Gold particles (1-μm diameter) were coated with 5 μg of the DNA as the instruction (Bio-Rad, Hercules, CA, USA), and then it was bombarded into onion epidermal cell at 1200 psi helium pressure utilizing a PDS-1000 system (Bio-Rad). After incubating 24 h in darkness, the onion cells were surveyed by confocal laser scanning microscope (Olympus, Japan). The 35S::GFP vector was as a reference.
Results
Cloning and characterization of the DcFT gene
The full-length cDNA of DcFT was obtained from carrot (D. carota) by producing the sequence using degenerate primers amplification and the RACE technology. The full-length cDNA of DcFT was 853 bp (GenBank accession number KY768910), comprising a 45-bp 5′-untranslated region (UTR), a 283-bp 3′-UTR, and an ORF 525-bp (Supplemental Fig. S1) encoding 175 amino acid residues with an estimated molecular weight of 20.0 kDa and a theoretical isoelectric point (PI) of 7.75.
According to the NCBI web server (https://www.ncbi.nlm.nih.gov/Structure/cdd/docs/cdd_search.html), the domain structure of the predicted amino acid sequence showed the DcFT was a member of the CENTRORADIALIS/TERMINAL FLOWER 1/SELF-PRUNING (CETS) gene family, which shares homology to and is alternatively called the PEBP gene family (Supplemental Fig. S2). An alignment of the DcFT protein and other FT protein is exhibited in Fig. 1. The amino acid Tyr88 (Y) and Gln144 (Q) belonging to conservative amino acid residues in FT-like proteins are important and are characteristic amino acids for distinguishing FT-like floral promoters (Hanzawa et al. 2005; Ahn et al. 2006). The amino acid sequences all contained the “Segment A domain (14AA)” and LYN, corresponding to a potential ligand-binding pocket in FT/TFL1 family proteins (Ahn et al. 2006). Capitalized letters on the alignment denote functionally vital conservative residues of FT proteins (Manoharan et al. 2016). A BLASTp comparison of the deduced amino acid sequence of DcFT with those of other FT proteins revealed that the protein of DcFT had the highest identify (88.00%) to that of CsFT and the lowest (62.22%) to that of AfFT (Supplement Table S2).
Alignment of the deduced amino acid sequence of DcFT and other homologous proteins in plants. A dark blue background represents amino acid 100% identity, and a light gray background represents amino acid identity > 75%. GenBank accession numbers of sequences in the figure are as follows: Vitis vinifera (VvFT, ABL98120.1), Gypsophila paniculata (GpFT, BAK23998.1), Gentiana triflora (GtFT, BAK40194.1), Arabidopsis thaliana (AtFT, AAF03936.1), Petunia × hybrida (PhFT, BAV21621.1), Camellia sinensis (CsFT, BAM83573.1), Beta vulgaris subsp. vulgaris (BvFT, AEI55782.1), Brassica oleracea (BoFT, ACH86033.1), and Daucus carota (DcFY, KY768910). The three rectangles in yellow represent conserved regions and amino acids shown in red indicate the Tyr85 (Y) and Gln140 (Q) residues that distinguish all FT-like members
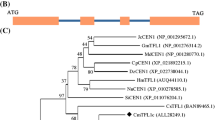
An alignment of the DcFT protein and 18 other plant FT proteins retrieved from NCBI was performed using ClustalW, and then an unrooted phylogenetic tree was constructed by the neighbor-joining method in the MEGA 6.0 software (Fig. 2). The phylogenetic tree indicated that DcFT protein had the closest relationship with GpFT and was only distantly related to FT from Vitis vinifera. The intimate relationship between DcFT and GpFT explains that they probably have similar functions in promoting flowering.
Phylogenetic tree of DcFT and other plant FT proteins. Numbers at branch points show bootstrap support. GenBank accession numbers of sequences in the figure are as follows: Paeonia suffruticosa (PsFT, AKS43552.1), Populus tomentosa (PtFT, AFU08240.1), Camellia sinensis (CsFT, BAM83573.1), Malus domestica (MdFT, ACL98164.1), Litchi chinensis (LcFT, AEU08965.1), Lactuca sativa (LsFT, AFV46423.1), Vitis vinifera (VvFT, ABL98120.1), Gypsophila paniculata (GpFT, BAK23998.1), Gentiana triflora (GtFT, BAK40194.1), Brassica oleracea (BoFT, ACH86033.1), Sinapis alba (SaFT, ACM69283.1), Arabidopsis thaliana (AtFT, AAF03936.1), Petunia × hybrida (PhFT, BAV21621.1), Nicotiana tabacum (NtFT, AGJ83935.1), Allium fistulosum (AfFT, AGU12402.1), Beta vulgaris subsp. vulgaris (BvFT, AEI55782.1), and Medicago truncatula (MtFT, XP_003624575.1)
Gene expression analysis of DcFT
Florigen, which produced in leaves and transmitted to the shoot apex, is primarily seduced by a systemic signal (Chailakhyan 1968; Mathieu et al. 2007). The expression pattern of numerous FT-like homologs has been analyzed, such as CsatFT1–CsatFT4 in perennial geophyte saffron crocus (Crocus sativus) (Tsaftaris et al. 2013). In order to elucidate the DcFT expression pattern in carrot, a qRT-PCR experiment was carried out to detect the transcript levels DcFT in various tissues/organs of carrot. In the complete floral organ stage, the expression level in rachises and petiole was the highest and lowest, respectively (Fig. 3a). Among the three different growth stages in carrot, the expression level of DcFT in leaves was the lowest in the third stage, stem and leaf was the highest in the obvious-meristem stage, and in the pre-anthesis (no flower primordium) stage, the expression level of DcFT in root and stem was approximately equality (Fig. 3b). After 10DAA, the expression level of DcFT in inflorescence sharply increased and then slowly increased reaching the maximum at 30DAA (Fig. 3c). In conclusion, these results illuminated that the expression of DcFT existed among tissues during developmental periods and was different, manifesting that DcFT was likely to play a central role in regulating flowering in carrot.
Expression patterns of DcFT in various tissues and organs; roots, stems, and leaves from three periods and inflorescences at different times. qRT-PCR was used to analyze the relative expression level of DcFT relative to β-actin protein as a control. Data are mean values and error bars represent the standard deviation from three biological replicates. 1–3 represent the pre-anthesis (no flower primordium), obvious-meristem (namely, the flower primordium), and complete floral organ stages (that is the flower unfolding)
Subcellular localization of DcFT
To determine the cellular location of the DcFT protein, the fusion protein expression vector 35S::DcFT-GFP was constructed by fusing the C-terminus of the DcFT protein to the green fluorescence protein (GFP) with the 35S promoter of the Cauliflower Mosaic Virus (CaMV). Then, the DNA plasmid of 35S::DcFT-GFP was guided into onion epidermal cells employing a gene gun. An expression vector 35S::GFP was as a reference. After cultivating for 24 h, 35S::DcFT-GFP and the empty 35S::GFP protein signal were detected by fluorescence microscopy, respectively (Fig. 4a–c, d–f). The fusion protein 35S::DcFT-GFP was clearly distributed in the nucleus and cytoplasm (Fig. 4d–f). This consequence was in line with the localization of NtFT protein from tobacco (Nicotiana tabacum L.) (Harig et al. 2012).
Subcellular localization of 35S::DcFT-GFP. a–c Subcellular localization of 35S::GFP; the panels represent fluorescence signal (excitation, 488 nm), bright field, and merged images, respectively. d–f Subcellular localization of 35S::DcFT-GFP; the panels represent fluorescence signal (excitation, 488 nm), bright field, and merged images, respectively
Discussion
Genetic and molecular analyses of various horticultural plants have indicated that FT homologs play a center role in regulating flowering time, for example, in Chinese Cymbidium , Sinapis alba , and Carya cathayensis (Huang et al. 2012; Chen et al. 2009), but the molecular mechanism of FT gene has not been studied in carrot. In this present research, isolating and cloning a homologous FT gene (DcFT) from carrot according to the conserved sequences of FT-like gene that was from various plants. The deduced protein sequence of DcFT was by BLAST search in the NCBI database indicating that DcFT proteins belong to the PEBP family and have the conserved domain sequences that promote flowering (Ahn et al. 2006). Furthermore, phylogenetic analysis distinctly revealed an intimate genetic relationship between DcFT and GpFT or VvFT. This conclusion was likely to suggest DcFT has similar functions to GpFT and VvFT, which participated in the control of flowering (Debener and Winkelmann 2010; Vergara et al. 2016).
The expression pattern of DcFT was very wide in various tissues but was predominantly expressed in inflorescences (30DAA) and rachises (Fig. 3a, c), which was consistent with the rest of FT homologs (Huang et al. 2012). In numerous plants, the expression patterns of FT-like homologs are evidently different in various tissues. In tobacco, NtFT1–NtFT4 were mostly expressed in leaves (Harig et al. 2012). In onion (Allium cepa L.), the expression level of FT genes (AcFT1-AcFT7) was detected in seed, leaf, bulb, and flowering bulb (Lee et al. 2013; Manoharan et al. 2016). In seedlings and mature period of A. thaliana, AtFT was expressed in every tissue, and especially, in flowers and immature siliques, the expression levels were higher (Kobayashi et al. 1999). In this study, the relative expression level of DcFT in leaf was the highest at the obvious-meristem (namely, flower pirmordium) stage and decreased immediately to basal levels at the complete floral organ stage (Fig. 3b), which may suggest that DcFT has a function in photosynthesis for regulating flowering time in carrot (Daucus carota L.). With the growth, the expression pattern of DcFT in inflorescences sharply increased and then slowly increased reaching the maximum at 30DAA (Fig. 3c), which suggest that the FT gene transfers to floral organ and promotes flowering with gradually accumulating after flowering. How the FT gene promotes flowering and the related mechanism need to further studied. Now, an experiment was under way involving transgenic tobacco plants, comparing short-day contition (8-h light/16-h dark) with long-day condition (16-h light/8-h dark) to study the mechanisms of FT gene intensively in carrot (Daucus carota L.).
Our data indicates that the protein of DcFT is localized in cytoplasm and nucleus (Fig. 4) which is in line with that of CsFT and FaFT (Higuchi et al. 2013; Lei et al. 2015), suggesting DcFT may play an important role in flowering regulation.
References
Abe M, Kobayashi Y, Yamamoto S, Daimon Y, Yamaguchi A, Ikeda Y et al (2005) Fd, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 309(5737):1052
Ahn JH, Miller D, Winter VJ, Banfield MJ, Lee JH, Yoo SY, Henz SR, Brady RL, Weigel D (2006) A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J 25:605–614
Ahn JH, Miller D, Winter VJ, Banfield MJ, Lee JH, Yoo SY, Henz SR, Brady RL, Andrés F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13:627–639
Bohlenius H, Huang T, Charbonnel-Campaa L, Brunner AM, Jansson S, Strauss SH, Nilsson O (2006) CO/FT regulatory module controls timing of flowering and seasonal growth cessation in trees. Science 312(5776):1040–1043
Boss PK, Bastow RM, Mylne JS, Dean C (2004) Multiple pathways in the decision to flower: enabling, promoting, and resetting. Plant Cell 16(Suppl):S18–S31. https://doi.org/10.1105/tpc.015958
Chailakhyan MK (1968) Internal factors of plant flowering. Annu Rev Plant Physiol 19(19):1–37
Chen FF, Huang YJ, Wang ZJ, Liu GH, Huang JQ (2009) Cloning and sequence analysis of the FLOWERING LOCUS T homologous genes from Carya cathayensis. Journal of Southwest Forestry University
Debener T, Winkelmann T (2010) Ornamentals. Biotechnol Agric For 64:369–391
Fukuda M, Matsuo S, Kikuchi K, Kawazu Y, Fujiyama R, Honda I (2011) Isolation and functional characterization of the FLOWERING LOCUS T homolog, the LsFT gene, in lettuce. J Plant Physiol 168:1602–1607
Guo D, Li C, Dong R, Li X, Xiao X, Huang X (2015) Molecular cloning and functional analysis of the FLOWERING LOCUS T (FT) homolog GhFT1 from Gossypium hirsutum. J Integr Plant Biol 57(6):522–533
Hanzawa Y, Money T, Bradley D (2005) A single amino acid converts a repressor to an activator of flowering. Proc Natl Acad Sci U S A 102:7748–7753
Harig L, Beinecke FA, Oltmanns J, Muth J, Müller O, Rüping B, Twyman RM, Fischer R, Prüfer D, Noll GA (2012) Proteins from the FLOWERING LOCUS T-like subclade of the PEBP family act antagonistically to regulate floral initiation in tobacco. Plant J 72:908–921
Higuchi Y, Narumi T, Oda A, Nakano Y, Sumitomo K, Fukai S et al (2013) The gated induction system of a systemic floral inhibitor, antiflorigen, determines obligate short-day flowering in chrysanthemums. Proc Natl Acad Sci U S A 110(42):17137–17142
Ho WW, Weigel D (2014) Structural features determining flower promoting activity of Arabidopsis FLOWERING LOCUS T. Plant Cell 26:552–564
Huang W, Fang Z, Zeng S, Zhang J, Wu K, Chen Z et al (2012) Molecular cloning and functional analysis of three FLOWERING LOCUS T (FT) homologous genes from Chinese cymbidium. Int J Mol Sci 13(9):11385
Kobayashi Y, Kaya H, Goto K, Iwabuchi M et al (1999) A pair of related genes with antagonistic roles in mediating flowering signals. Science 286:1960–1962
Komeda Y (2004) Genetic regulation of time to flower in Arabidopsis thaliana. Annu Rev Plant Biol 55:521–535. https://doi.org/10.1146/ annurev.arplant.55.031903.141644
Lee R, Baldwin S, Kenel F, McCallum J, Macknight R (2013) FLOWERING LOCUS T genes control onion bulb formation and flowering. Nat Commun 4:2884
Lei H, Guo X, Wang Y, Yao L, Wang S, Li T (2015) Identification and characterization of faft1: a homolog of flowering locus t from strawberry. Adv J Food Sci Technol 8(3):180–188
Lifschitz E, Eviatar T, Rozman A, Shalit A, Goldshmidt A, Amsellem Z, Alvarez JP, Eshed Y (2006) The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc Natl Acad Sci U S A 103:6398–6403
Lin MK, Belanger H, Lee YJ, Varkonyi-Gasic E, Taoka K, Miura E, Xoconostle-Cázares B, Gendler K, Jorgensen RA, Phinney B, Lough TJ, Lucas WJ (2007) FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell 19:1488–1506
Manoharan RK, Han JS, Vijayakumar H, Subramani B, Thamilarasan SK, Park JI et al (2016) Molecular and functional characterization of FLOWERING LOCUS T homologs in Allium cepa. Molecules 21(2)
Mathieu J, Warthmann N, Küttner F, Schmid M (2007) Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr Biol 17:1055–1060
Niu L, Fu C, Lin H, Wolabu TW, Wu Y, Wang ZY et al (2016) Control of floral transition in the bioenergy crop switchgrass. Plant Cell Environ 39:2158–2171
Pin PA, Benlloch R, Bonnet D, Wremerth-Weich E, Kraft T, Gielen JJL et al (2010) An antagonistic pair of FT homologs mediates the control of flowering time in sugar beet. Science 330:1397–1400
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Samach AH, Onouchi SE, Gold GS, Ditta Z, Schwarz-Sommer et al (2000) Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616
Schmittgen TD, Livak KJ (2008) Schmittgen td, livak kjanalyzing real-time PCR data by the comparative c(t) method. Nat Protoc 3(6):1101–1108
Scott A, Wyatt S, Tsou PL, Robertson D, Allen NS (1999) Model system for plant cell biology: GFP imaging in living onion epidermal cells. BioTechniques 26(6):1125–1132
Srikanth A, Schmid M (2011) Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci 68:2013–2037
Sun H, Jia Z, Cao D, Jiang B et al (2011) GmFT2a, a soybean homolog of FLOWERING LOCUS T, is involved in flowering transition and maintenance. PLoS One 6(12):e29238
Tamaki S, Matsuo S, Wong HL, Yokoi S et al (2007) Hd3a protein is a mobile flowering signal in rice. Science 316:1033–1036
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) Mega6: molecular evolutionary genetics analysis version 6.0. Molecular Biology & Evolution 30(12):2725
Tan FC, Swain SM (2006) Genetics of flower initiation and development in annual and perennial plants. Physiol Plant 128:8–17
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. https://doi.org/10.1093/nar/22.22.4673
Tian C, Jiang Q, Wang F, Wang GL, ZS X, Xiong AS (2015) Selection of suitable reference genes for qPCR normalization under abiotic stresses and hormone stimuli in carrot leaves. PLoS One 10(2):e0117569
Tsaftaris A, Pasentsis K, Argiriou A (2013) Cloning and characterization of flowering locus t-like genes from the perennial geophyte saffron crocus (Crocus sativus). Plant Mol Biol Report 31(6):1558–1568
Turck F, Fornara F, Coupland G (2008) Regulation and identity of florigen: FLOWERING LOCUS T moves center stage. Annu Rev Plant Biol 59(59):573–594
Vergara R, Noriega X, Parada F, Dantas D, Pérez FJ (2016) Relationship between endodormancy, FLOWERING LOCUS T, and cell cycle genes in vitis, vinifera. Planta 243(2):411–419
Wigge PA, Kim MC, Jaeger KE, Busch W, Schmid M et al (2005) Integration of spatial and temporal information during floral induction in Arabidopsis. Science 309:1056–1059
Wilkie JD, Sedgley M, Olesen T (2008) Regulation of floral initiation in horticultural trees. J Exp Bot 59:3215–3228
Wolabu TW, Zhang F, Niu L, Kalve S, Bhatnagar-Mathur P, Muszynski MG et al (2016) Three FLOWERING LOCUS T-like genes function as potential florigens and mediate photoperiod response in sorghum. New Phytol 210(3):946
Zeevaart JA (2008) Leaf-produced floral signals. Curr Opin Plant Biol 11(5):541–547
Funding
This work was funded by grants from the National Key Technology Research and Development Program of China (2012BAD01B00, 2012BAD50G01, 2014BAD01B08, and JNKYT201601). Our research was also supported by the Technological Innovation Capacity Program of the Beijing Academy of Agricultural and Forestry Sciences (KJCX20150111, KJCX20170102).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The experiments described here comply with the current laws of China.
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Zhan, Z., Zhang, C., Zhang, H. et al. Molecular cloning, expression analysis, and subcellular localization of FLOWERING LOCUS T (FT) in carrot (Daucus carota L.). Mol Breeding 37, 149 (2017). https://doi.org/10.1007/s11032-017-0749-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-017-0749-y