Abstract
A genetic linkage map has been constructed from an interspecific diploid blueberry population [(Vaccinium darrowii Fla4B × Vaccinium corymbosum W85-20) F1#10 × V. corymbosum W85-23] designed to segregate for cold hardiness and chilling requirement. The map is comprised of 12 linkage groups (equivalent to the haploid chromosome number of diploid blueberry) and totals 1,740 cM. Included on the map are 265 markers based on simple sequence repeats, expressed sequence tag-polymerase chain reactions, single nucleotide polymorphisms, and randomly amplified polymorphic DNAs. The estimated map coverage is 89.9 %, and the average distance between markers is 7.2 cM. The mapping population was evaluated for 2 years (2009 and 2010) for mid-winter bud cold hardiness and for 3 years (2011–2013) for chilling requirement under controlled conditions. Broad-sense heritability of both cold hardiness and chilling requirement was quite high under these conditions with values of 0.88 and 0.86, respectively. One quantitative trait locus/loci (QTL) for cold hardiness and one for chilling requirement were identified that were consistent over at least 2 years. A second weaker QTL for chilling requirement was detected in only one of the 3 years.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Blueberry production has risen by about 50 % over the last 5 years, making it one of the most rapidly expanding fruit crops in the world today. Popularity among consumers has increased, too, no doubt propelled by research findings on their many health benefits. For example, blueberries have been shown to be an especially rich source of anthocyanins (Prior et al. 1998), which have been associated with many health benefits from reducing eyestrain, improving night vision, and helping prevent macular degeneration, to exhibiting anticancer activity (Cho et al. 2004; Kalt et al. 2007). A recent human study found that blueberries may even improve certain features of metabolic syndrome and related cardiovascular risk factors like high blood pressure at dietary achievable doses (Basu et al. 2010).
Blueberry is a native North American fruit crop. Several species of various ploidy levels within the Cyanoccocus section of the genus Vaccinium, family Ericaceae, are grown commercially. The major commercial types in the USA are cultivars of Vaccinium corymbosum L. (tetraploid highbush blueberry) and, to a lesser extent, V. virgatum Ait. (hexaploid rabbiteye blueberry; syn. V. ashei Reade), and native, managed stands of V. angustifolium Ait. (tetraploid lowbush blueberry). Highbush blueberry cultivars are classified into northern and southern types. Southern highbush cultivars, with low chilling requirements, have been developed through the introgression of the low-chilling southern diploid species Vaccinium darrowii Camp, through the production of unreduced gametes, into the tetraploid V. corymbosum background (Rowland et al. 2011). The southern highbush cultivars have the advantage of being earlier ripening than rabbiteye blueberries, which are also grown in the southern USA.
The highbush blueberry was domesticated in the USA in the early twentieth century. Breeding efforts have focused on the development of cultivars with broader climatic adaptation (increased freezing tolerance for northern regions and reduced chilling requirements for southern regions), season extension, disease and pest resistance, mechanical handling tolerance, and high fruit quality (Galletta and Ballington 1996). However, evaluating breeding populations in the field for traits related to climatic adaptation can be very difficult, as chilling hours and temperatures vary greatly from year to year and site to site. The availability of molecular markers for help in combining traits for climatic adaptation with other important traits like fruit and nutritional quality would be very useful. Woody perennials, like blueberry, are especially suitable for improvement via marker-assisted breeding because of their long generation times, high heterozygosity, self-infertility, inbreeding depression, and polyploidy of commercial types.
A study of winter injury in field-grown blueberry found heritability estimates over 3 years to be low although individual year estimates were higher. General combining ability variance components were consistently higher than specific combining ability, suggesting that additive variance is more important than nonadditive variance (Fear et al. 1985). However, because winter survival in the field is determined by multiple factors, the authors recommended screening for cold hardiness under controlled conditions to accurately ascertain additive, dominance, and epistatic interactions.
We have developed an interspecific diploid blueberry population suitable for mapping cold hardiness, chilling requirement, and multiple fruit quality traits (Rowland et al. 1999). This population is a testcross generated by crossing an F1 hybrid of a low-chilling, freezing-sensitive V. darrowii selection and a high-chilling, freezing tolerant diploid V. corymbosum selection, to another diploid V. corymbosum selection. A true F2 or backcross population would be difficult, if not impossible, to generate for mapping due to self-sterility (Galletta and Ballington 1996) and inbreeding depression (Krebs and Hancock 1988) in diploid blueberry. The testcross population described here, along with other related populations, has been used previously in genetic analyses of cold hardiness (Arora et al. 2000; Rowland et al. 1999) and chilling requirement (Rowland et al. 1998, 1999). Degree of dominance and generation means analysis revealed that cold hardiness is a quantitative trait controlled largely by additive gene effects and, to a lesser extent, by dominance gene effects, with cold sensitivity appearing to be partially dominant. Cold tolerance could be explained adequately by a simple additive-dominance model of gene action, although two epistatic models involving additive–additive and dominance–dominance interactions also fit the data (Arora et al. 2000). Formal generation means analysis has not been carried out successfully for chilling requirement; however, segregation patterns suggest that it, too, is largely quantitatively inherited with low chilling requirement showing additive and perhaps some dominance gene action (Rowland et al. 1998, 1999, 2011).
Here, the construction of a genetic linkage map for the interspecific diploid blueberry population described above is described. The map is comprised of 265 molecular markers and includes simple sequence repeat (SSR), expressed sequence tag-polymerase chain reaction (EST-PCR), single nucleotide polymorphism (SNP), and randomly amplified polymorphic DNA (RAPD) markers. We also report the identification of quantitative trait loci (QTL) for chilling requirement and cold hardiness by evaluating the population for these traits over 5 years under controlled conditions.
Materials and methods
Plant material
A blueberry interspecific diploid testcross population, segregating for mid-winter cold tolerance, chilling requirement, and fruit quality traits, was generated for genetic linkage map construction and QTL identification. The population, developed by crossing an interspecific F1 plant [V. darrowii selection Fla4B (low chilling, cold sensitive, evergreen) × diploid V. corymbosum selection W85-20 (high chilling, cold hardy, deciduous)] to another diploid V. corymbosum W85-23 (high chilling, cold hardy, deciduous, like W85-20), was propagated to give approximately three to four clones of each individual. The plants from this cross (F1#10 × W85-23) are maintained in a greenhouse in 4- to 12-l pots at the Beltsville Agricultural Research Center-West, Beltsville, MD. The selection Fla4B was originally collected from Florida, and W85-20 and W85-23 were collected from New Jersey (Arora et al. 2000). Fla4B has been used extensively in blueberry breeding programs and is still the primary source of low chilling requirement alleles in southern highbush cultivars (Rowland et al. 2011).
DNA extraction and general overview of marker scoring
DNA was extracted from young leaf tissue of 95 mature individuals of the mapping population (F1#10 × W85-23), parents, and grandparents as described previously (Rowland et al. 2003b) using a modified CTAB procedure. A variety of markers, including those based on EST-PCRs (Rowland et al. 2003b), SSRs, SNPs, and RAPDs, were generated and screened first on the parents (F1#10 and W85-23) and the grandparents (Fla4B and W85-20). Our intent was to develop a genetic map of the F1#10, and not W85-23, because of concerns that W85-23 may be fixed for genes responsible for phenological traits like cold hardiness and chilling requirement, and thus, these genes may not segregate in the mapping population. The F1#10, on the other hand, was specifically developed to be heterozygous for these genes. Therefore, only markers that met the following criteria were used for map construction: the marker must have been (1) polymorphic between Fla4B and W85-20, (2) present in the F1#10, and (3) absent in W85-23. Markers were scored on an EXCEL spreadsheet as present or absent in each individual.
Marker design and application
For the EST-PCR markers, emphasis was initially placed on designing primers from blueberry ESTs representing cold-responsive genes, identified previously from microarrays (Dhanaraj et al. 2007) and subtracted/reverse subtracted libraries (Naik et al. 2007). These ESTs were derived from Sanger sequencing and are present in the EST database of GenBank/NCBI (Dhanaraj et al. 2004, 2007). Later, PCR primers were designed from contigs assembled from transcriptome data obtained using 454 sequencing (Rowland et al. 2012). These 454 sequences are available on our Web site (http://bioinformatics.towson.edu/BBGD454/) and in the sequence read archive (SRA) of GenBank (under accessions SRX100856, SRX100859, and SRX100861-SRX100867). The EST-PCR marker names, forward and reverse primer sequences, origin of sequences, GenBank accession numbers, and the genes encoded by the sequences are given in Sheet 1 of the Online Resource 1 spreadsheet. Prefixes in marker names indicate the source of the ESTs from which the primers were designed. Methods for designing EST-PCR primers, amplifying products, and separating the products on agarose gels were those described in Rowland et al. (2003b).
SSR markers were developed from blueberry sequences originating from several different sources including: 454-sequenced ESTs provided at http://bioinformatics.towson.edu/BBGD454/ and in the SRA of GenBank (VCB prefix in marker names stands for V. corymbosum from ‘Bluecrop,’ Rowland et al. 2012); Sanger-sequenced ESTs in the EST database of GenBank (GVC prefix for GenBank-mined V. corymbosum); private Sanger-sequenced ESTs developed by the New Zealand Institute for Plant and Food Research Ltd. (GenBank accession numbers used for prefix); genomic sequences from the largest scaffolds of the current whole genome assembly of diploid blueberry selection W85-20 (KAN prefix for Kannapolis, NC, where the genome sequence is being assembled); and from sequences near specific gene sequences of interest identified from the whole genome assembly (various prefixes for gene names). The SSR marker names, forward and reverse primer sequences, and origin of sequences are given in Sheet 2 of the Online Resource 1 spreadsheet. Sequences were mined for SSRs and primers designed using the online SSR tool (SSR Server) available at the Genome Database for Vaccinium (GDV, http://www.vaccinium.org) as described in Rowland et al. (2012). Sanger ESTs from GenBank were mined after eliminating sequences that contained previously developed SSRs (Boches et al. 2006).
To identify SSRs near genes of interest, such as anthocyanin biosynthesis genes (Jaakola et al. 2002) and CBF (C-repeat or CRT binding factor), a transcriptional activator involved in cold acclimation (Jaglo-Ottosen et al. 1998; Liu et al. 1998), amino acid sequences of these genes were first identified from blueberry, grape, strawberry, or Arabidopsis, from the UniProt database. Sequences were then used in tBLASTn searches of the diploid V. corymbosum W85-20 genome assembly. Blueberry scaffolds with the best E values (threshold of e−10) were mined using the online Vaccinium SSR tool. Primer pairs for the best three SSRs were synthesized for screening. Criteria for best SSRs included: presence of di- or trinucleotide motifs close to the alignment’s starting location, large number of repeats, and expected PCR products of 100–700 bp.
PCR and capillary electrophoresis conditions for SSR genotyping varied somewhat among the laboratories that performed the work, depending on the equipment that was available and optimization protocols established for that equipment at each location. At the USDA-ARS, National Clonal Germplasm Repository laboratory (NCGR), PCR reactions contained 1× reaction buffer, 2 mM MgCl2, 0.2 mM dNTPs, 0.5 μM of fluorescent M13 primer (Schuelke 2000), 0.12 μM forward primer, 0.50 μM reverse primer, 0.075 units of GoTaq® DNA Polymerase (Promega Corporation, Madison, WI, USA), and 4.5 ng genomic DNA in a total volume of 15 μL. Amplification conditions consisted of 94 °C for 3 min; followed by 10 cycles of 94 °C for 40 s, 65 °C for 45 s (−1.0 °C per cycle), and 72 °C for 45 s; 20 cycles of 94 °C for 40 s, 52 °C for 45 s, and 72 °C for 45 s; eight cycles of 94 °C for 40 s, 53 °C for 45 s, and 72 °C for 45 s; and finally 72 °C for 30 min. Successful amplification was first assessed by 2 % agarose gel electrophoresis. PCR products were separated by capillary electrophoresis using a Beckman Coulter (Indianapolis, IN, USA) CEQ 8000 genetic analyzer and analyzed with software provided by the manufacturer.
At the New Zealand Institute for Plant and Food Research Ltd., PCR amplification contents were the same as those outlined for NCGR using the M13 fluorescently labeled approach of Schuelke (2000). Amplification conditions consisted of 94 °C for 2 min 45 s; then 10 cycles at 94 °C for 55 s, 60 °C for 55 s (−0.5 °C per cycle), and 72 °C for 1 min 30 s; 30 cycles at 94 °C for 55 s, 55 °C for 55 s, and 72 °C for 1 min 30 s; and finally 72 °C for 10 min. PCR products were analyzed on an ABI PRISM® 377 DNA sequence analyzer (Applied Biosystems, Foster City, CA, USA) using dye-labeled GeneScan™ -350 ROX™ size standard (Applied Biosystems) on Long Ranger® Singel® packs (Lonza Rockland, ME, USA), and a 64-well casting comb (0.18 mm; single use; no. 4309455 Shark’s Tooth Comb; Applied Biosystems) according to the manufacturer’s protocol. Allele sizes were visualized using Peak Scanner™ Software v.1.0 (Applied Biosystems).
At the USDA-ARS, Genetic Improvement of Fruits and Vegetables laboratory, the PCR conditions were the same as those at NCGR except that no fluorescent M13 primer was used. Separation of PCR products was achieved using the Advanced Analytical FS96 system and software provided by Advanced Analytical (Ames, IA, USA).
At the James Hutton Institute, PCR amplifications consisted of 25 μl reactions containing 1× PCR buffer (10 mm Tris–HCl, 1.5 mm MgCl2, 50 mm KCl, pH 8.3), 0.2 mM dNTPs, 1.0 μM fluorescently labeled forward primer, 1.0 μM reverse primer, 0.1 units Taq polymerase (Roche Applied Science, Indianapolis, IN, USA), and 25 ng template DNA. Amplification conditions were as follows: 95 °C for 5 min; followed by 40 cycles of 95 °C for 30 s, 57 °C for 30 s, and 72 °C for 45 s; followed by a final extension of 72 °C for 10 min. PCR products were first resolved on 1.5 % agarose gels followed by analysis on an ABI 3730 DNA sequence analyzer (Applied Biosystems) using Rox 500 (Applied Biosystems) as an internal size standard. Allele sizes were visualized using GeneMapper software version 3.7 (Applied Biosystems).
Some of the monomorphic SSR markers were screened using the high-resolution melting (HRM) point analysis technique, in an attempt to detect SNPs and other small sequence and length variations between different PCR products (Reed and Wittwer 2004; Wittwer et al. 2003; Montgomery et al. 2007). HRM-PCRs and analyses were performed as described in Bushakra et al. (2012), with conditions as outlined in Chagne et al. (2008). Names of the mapped SNP markers (based on HRM), forward and reverse primer sequences, and origin of sequences are shown in Sheet 3 of the Online Resource 1 spreadsheet.
RAPDs were generated using 10-base RAPD primers from Operon Technologies, Inc. (Alameda, CA, USA) and from the Biotechnology Laboratory, University of British Columbia (Vancouver, British Columbia, Canada). DNA amplification and electrophoresis conditions were those described in Rowland et al. (2003b) except that 0.2 μM of primer was used. Primers that generated mapped markers are listed in Sheet 4 of the Online Resource 1 spreadsheet.
Map construction
Markers that met mapping criteria described above were used for map construction. A linkage map for the parent F1#10 was generated using the regression approach in JoinMap 4.0 (Van Ooijen 2006). Linkage groups (LG) were established with independence LOD of 4.0–5.0; and Haldane’s mapping function was used to calculate map distances. Other default parameters of JoinMap were accepted. Several methods were used to minimize potential sources of error in the map including removing markers that had the most skewed segregation ratios (P < 0.0001), appeared repeatedly in genotypes with very low probabilities in the double recombinant table, were the least informative, and had suspicious linkages. The graphical presentation of the map was prepared with MapChart 2.2 (Voorrips 2002). Map coverage was calculated by method four of Chakravarti et al. (1991).
Cold hardiness determinations
To achieve cold acclimation, the mapping population, along with parents and grandparents, were moved from a heated to an unheated hoop house at the USDA-ARS in Beltsville, MD, in late December and early January of 2008–2009 and 2009–2010. Plants were evaluated for cold hardiness using a detached shoot assay as previously described (Arora et al. 2000, 2004; Rowland et al. 2005) when they had received approximately 500–550 chill units (hours of exposure to temperatures in the range of 0–7 °C, measured as described below under ‘chilling requirement evaluations’). Hoop house temperatures were monitored with a biophenometer, Model TA51 (Omnidata International, Inc., Logan, UT, USA). Beginning in early February of 2009 and 2010, ~35 terminal shoots were collected from several plants (1–4) of each genotype. Sets of shoots, 5 to 6 cm long with at least three flower buds attached, were subjected to a freeze–thaw protocol, which included exposure to a range of temperature treatments. The three most distal buds on each of three shoots were evaluated for damage per treatment temperature.
The protocol consisted of placing three randomly sampled shoots/treatment temperature from each genotype in test tubes with 0.5 ml of water and freezing them in a glycol bath (model 2325; Forma Scientific, Marietta, OH, USA). Bud temperature was monitored by copper-constantan thermocouples (TT-T-30) attached to a thermometer (DP465, Omega Engineering, Stamford, CT, USA). Ice nucleation was initiated at −1 °C. Samples were allowed to equilibrate for 45 min and then further cooled at 0.5 °C/30 min down to −4 °C, at 1 °C/30 min down to −8 °C, and at 2 °C/30 min thereafter to respective final treatment temperatures. Treatment temperatures chosen for cold-acclimated buds spanned a range from −10 to −28 °C at 2 °C increments, to represent 0–100 % injury for most, if not all, genotypes. Controls consisted of shoots held at 4 °C without exposure to freezing. To assess injury, shoots were removed from the freezing bath at respective treatment temperatures and samples were allowed to thaw overnight at 4 °C followed by a 24-h incubation at 20 °C. Subsequently, the three most distal buds were dissected and observed for injury (visual browning) of the ovaries in individual flowers (Arora et al. 2000; Flinn and Ashworth 1994), and a percent damage value was calculated for each bud.
Bud cold hardiness was measured as LT50 values, i.e., the temperature causing 50 % injury overall. For each genotype × year combination, bootstrap estimates (Manly 1997) of LT50 values and their 95 % fiducial confidence intervals were calculated using the PROBIT procedure of the Statistical Analysis System (SAS) 9.2 package (SAS Institute Inc., Cary, NC, USA). For this, the nine observed data points (three distal buds on each of the three shoots) for each temperature were resampled (n = 9 with replacement) 30 times. For each genotype, year and replicate, the 30 LT50 estimates were used to fit a logistic regression model, from which an LT50 and a 95 % confidence interval were estimated.
Chilling requirement determinations
The mapping population, as well as parents and grandparents, were evaluated for chilling requirement at the USDA-ARS in Beltsville, MD, USA, in the winters of 2010–2011, 2011–2012, and 2012–2013. One to six plants of each genotype were held in a heated greenhouse until November of each year, specifically until 11-26-10, 11-25-11, and 11-9-12, at which point plants had set a sufficient number of flower buds and were dormant. On those dates, plants were moved into an unheated hoop house covered with opaque winter-white plastic to minimize daytime temperature increases and were exposed to ambient light and temperature conditions. Chill units (hours from 0 to 7 °C) were measured with a Lascar EL-USB-2-LCD+ datalogger (Lascar Electronics, Erie, PA, USA). Sets of shoots (2–6), 5 to 6 cm long with three to eight flower buds attached, from multiple plants of each genotype, were collected every 200 chill units (200, 400, 600, 800, and 1,000). Shoots were then rated for floral bud development after 3 weeks at 24 °C (14-h photoperiod) as described by Spiers (1978). Accordingly, each bud was examined for signs of budbreak and given a score from stage 1 through 7, with 1 representing the stage with no sign of visible swelling, 2 representing the stage of first visible swelling and separation of bud scales, etc., and finally 7 representing the stage after which corollas had completely expanded and dropped. Mean chilling requirements were determined from the percentage of breaking buds (growth stage >2) after plants had received the various chill units (200, 400, 600, 800, and 1,000) as described in Rowland et al. (1998). Mean chilling requirements were calculated by fitting a straight-line regression equation of the frequency of buds at growth stage >2 against the number of chill units. Slopes and y-intercepts of equations with r 2 values >0.75 were used to calculate the chill units required for 50 % of the buds to break (CR50).
Distribution, correlation, and heritability analyses
The normality of cold hardiness and chilling requirement distributions was assessed using the UNIVARIATE procedure of SAS v. 9.3. The CORR procedure was used to test for correlations between phenotypic data from different years for the same trait and between different traits. The Pearson correlation coefficient (r) was used, because both traits had a normal distribution pattern most years. Correlations were considered significant at P < 0.001. Correlations were classified as high if r > 0.7, moderate if 0.4 < r < 0.7, and weak if r < 0.4.
Broad-sense heritability (H) was estimated for cold hardiness and chilling requirement as the ratio of genotypic variance to total phenotypic variance. SAS PROC MIXED (SAS v. 9.2) was used to determine variance components for
where r = number of replications and e = number of environments (years) (Holland et al. 2003). Mean squares (genotype and genotype × environment) from PROC GLM (SAS v. 9.2) were used to construct the 95 % confidence intervals (Knapp et al. 1985).
QTL analyses
Phenotypic data for chilling requirement (CR50) and cold hardiness (LT50) were used in QTL analyses with MapQTL 5.0 (Van Ooijen 2004). Because a map was not developed for the parent W85-23, however, these analyses are limited to markers and QTL present in the F1#10 that contribute to the observed phenotypic variation. The genome-wide (GW) thresholds for each trait and year were calculated with the permutation test (PM) set at 1,000 permutations. The QTL were considered significant in interval mapping (IM) and restricted multiple QTL mapping (rMQM) when the LOD score exceeded the GW 90th percentile (i.e., significant at 10 % error rate). Forward selection was performed by first identifying QTL by IM, selecting nearest markers to set as cofactors, re-analyzing with rMQM, and then repeating rounds of cofactor selection and rMQM until no additional QTL were identified.
Results
Plant population chosen for mapping
The generation of interspecific diploid testcross populations that segregate for mid-winter cold tolerance and chilling requirement has been described in detail previously (Rowland et al. 1999). Briefly, three V. darrowii selection Fla4B (low chilling, cold sensitive, and evergreen) × V. corymbosum selection W85-20 (high chilling, cold hardy, and deciduous) hybrids were crossed to another V. darrowii NJ88-13-15 and another V. corymbosum W85-23. Fla4B has an average mid-winter cold tolerance LT50 of −14.5 °C and chilling requirement of 330 chill units. W85-20 has a cold tolerance LT50 of −25.5 °C and chilling requirement of 790 chill units. The various populations were evaluated for chilling requirement and cold tolerance to identify the best one to use for the identification of QTL (data not shown). Based on the results, we decided to focus on the cross of F1#10 × W85-23, which had a wide distribution of both chilling requirement and cold hardiness levels (Fig. 1).
Distribution of cold hardiness and chilling requirement in the diploid blueberry mapping population. a Distribution of cold hardiness (LT 50 in °C) in the diploid blueberry mapping population [(Fla4B × W85-20) F1#10 × W85-23] in 2009 and 2010. Cold hardiness levels of parent plants are indicated by arrows. b Distribution of chilling requirement (chill units required for 50 % bud break) in the diploid blueberry mapping population in 2011, 2012, and 2013. Chilling requirements of parent plants are indicated by arrows
Phenotypic data
The distribution of cold hardiness in the mapping population for the 2 years tested, 2009 and 2010, was normal according to the Shapiro–Wilk and Kolmogorov–Smirnov tests for normality (Fig. 1a). The cold hardiness levels for the 2 years were moderately correlated (r = 0.663; P < 0.0001) (Table 1). The distribution of chilling requirement for the 3 years tested, 2011, 2012, and 2013, was also normal except for the 2013 data (according to the Shapiro–Wilk test), where the distribution was somewhat skewed toward a preponderance of genotypes with relatively low chilling requirements (Fig. 1b). The chilling requirements for the 3 years were moderately to highly correlated (for CR2011:CR2012, r = 0.587, P < 0.0001; for CR2012:CR2013, r = 0.694, P < 0.0001; for CR2011:CR2013, r = 0.763, P < 0.0001) (Table 1). Cold hardiness appeared weakly negatively correlated with chilling requirement but only for the CH2009:CR2012 comparison (the lower the cold hardiness LT50, the higher the chilling requirement). This was only significant at the P < 0.01 level (Table 1).
Broad-sense heritability (H) for cold hardiness was estimated to be 0.88, with a 95 % confidence interval of 0.70–0.88. For chilling requirement, H was calculated to be 0.86, with a 95 % confidence interval of 0.78–0.90. Because of the way chilling requirement was determined, however, years were treated as reps, and there were no within-year reps. Therefore, in the H calculation, there was no genotype × environment component, and hence, H may be overestimated for chilling requirement if the genotype × environment interaction is large.
Map construction
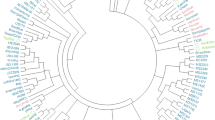
Blueberry progeny from the cross of F1#10 × W85-23 was genotyped for 340 markers. Of these, 265 markers were mapped (Fig. 2) and are described in detail in the Online Resource 1 spreadsheet. In total, 219 primer pairs generated the 265 mapped markers. A summary of the types of markers mapped, the percentage showing distorted segregation ratios, the percentage that yielded multiple mapped fragments, and, of those, the percentage that were allelic, is shown in Table 2. Most of the 265 mapped markers segregated 1:1, as expected for dominant markers in a testcross population (Aa × aa). However, 74 of the markers still had significantly distorted ratios from the expected 1:1 ratio (P < 0.05) even after removal of those with the most skewed segregation ratios (P < 0.0001). The markers with significantly distorted ratios are indicated by asterisks on Sheet 5 of the Online Resource 1 spreadsheet along with their positions on the map.
Genetic linkage map of diploid blueberry population [(Fla4B × W85-20) F1#10 × W85-23]. Markers are described in detail in Online Resource 1. Mapped markers include EST-PCR, SSR, SNP, and RAPD markers (Table 2). Location of QTL for cold hardiness (CH) and chilling requirement (CR) is indicated to the right of the linkage groups by different patterned bars. The bars represent 2-LOD (logarithm of the odds) support intervals based on the results from rMQM (restricted multiple QTL mapping)
The 265 mapped markers were organized into 12 LG (Fig. 2), which is equal to the haploid chromosome number for diploid blueberry. The total map length was 1,740 cM, with an average distance between markers of 7.2 cM. The largest gap was 33 cM. LG ranged in size from 56 to 220 cM, and the number of markers per LG ranged from 5 to 50. The average number of markers per LG was 22.1. The estimated map coverage, calculated according to method four of Chakravarti et al. (1991), was 89.9 %.
QTL analyses
We identified one QTL for cold hardiness and two for chilling requirement from the F1#10 genetic map (Fig. 2) and 5 years of data (2 years for cold hardiness and 3 years for chilling requirement evaluations). Table 3 provides detailed information (LOD scores, % trait variance explained, etc.) for the QTL and their associated markers each year.
In both 2009 and 2010, one QTL for cold hardiness was identified on LG 4 by IM followed by restricted multiple QTL mapping (rMQM). The nearest marker to the QTL peak was EST-PCR marker leaf-00248. This QTL was significant at the 0.05 level in 2009 (LOD 5.12, GW95 % 3.1) and in 2010 (LOD 3.14, GW95 % 3.1). Percentage of phenotypic variance explained by the cold hardiness QTL was 32.9 and 20.1, in 2009 and 2010, respectively.
Chilling requirement QTL were identified in 2 of the 3 years. IM followed by rMQM analysis identified a QTL on LG 6 that coincided with the position of the RAPD marker OPP-09b in 2012 (LOD 3.31, GW95 % 3.3) and in 2013 (LOD 3.5, GW95 % 3.0). With OPP-09b used as a cofactor in rMQM, a second QTL was identified near RAPD marker OPP-14d on LG 8 only in 2013 (LOD 2.84, GW90 % 2.7). The LG 6 QTL explained 18.6 and 16.7 % of the phenotypic variance in 2012 and 2013, respectively, while the LG 8 QTL from 2013 explained 14.6 %. In 2011, marker OPP-09b on LG 6 still contributed to the phenotypic variance (10.8 % explained) although the LOD score was below the threshold for 90 % GW significance.
Discussion
Here, we have described the first genetic linkage map of diploid blueberry based primarily on SSR and EST-PCR markers, and of sufficient density for QTL mapping. Relatively, low-density maps of diploid (Rowland and Levi 1994; Rowland et al. 1999, 2003a) and tetraploid blueberry (Qu and Hancock 1997) have been published previously but were comprised mainly of RAPD markers. An earlier map of a related V. darrowii × V. corymbosum hybrid population (based on several subpopulations), composed of only about 100 RAPD markers (Rowland et al. 1999, 2003a), was used to identify one putative QTL for cold hardiness (Rowland et al. 2003a).
Here, blueberry progeny from a single population (F1#10 × W85-23) was genotyped for over 300 total markers. Of these, 265 markers were mapped to 12 LG, equivalent to the haploid chromosome number for diploid blueberry. A couple of the LG were quite short, however, with only 5–8 markers. Perhaps coverage of these LG was lacking because, so far, most of the mapped markers were derived from expressed genes (EST-PCRs and EST-SSRs). An effort to map more genomic SSRs may remedy this problem in the future. About 28 % of the mapped markers (74/265) had significantly distorted segregation ratios, even though 17 with the most highly distorted ratios (significant at P < 0.0001) had been removed before the analysis. Considering that F1#10 is an interspecific hybrid, the percentage of markers with significantly distorted ratios was still not particularly high. Of the mapped markers with distorted ratios, 24 were clustered on LG 1 in a region between 67 and 144 cM, 13 clustered on LG 3 between 44 and 88 cM, and 5 clustered on LG 5 between 109 and 173 cM, suggesting these may be of biological significance.
A map of cranberry (V. macrocarpon), which is closely related to blueberry, consisting of 136 distinct loci was recently published (Georgi et al. 2013). The map is based on SSR markers, derived from blueberry ESTs and from cranberry genomic sequences, and cranberry-derived sequence-characterized amplified region (SCAR) markers. Some of the markers were designed from SSRs near sequences of interest based on the draft cranberry genome assembly, such as flavonoid biosynthetic pathway genes and defense-related genes. Markers in common between the blueberry and cranberry maps are shown in the Online Resource 2 table. Even though the cranberry map utilized 33 SSR primer pairs from blueberry, only four of these primer pairs gave markers that were also mapped in blueberry. In addition, markers generated from SSR primer sequences located near a chalcone isomerase gene were mapped in both blueberry and cranberry; the SSR primer sequences were different, however. With so few markers in common and the whole genome assemblies for blueberry and cranberry still unpublished, little can be compared between the two maps at this point. None of the shared markers were linked on the blueberry map except for the pair of fragments, CA325_205 and CA325_227, and the pair, CHI03186-1_124 and CHI03186-1_128, which mapped to LG 11. In cranberry, the comparable markers, CA325 and Scf12916, mapped to LG 8, suggesting that the blueberry LG 11 corresponds to the cranberry LG 8. Another pair of shared markers, GVC-V24d10b_202 and 2ms4d10b, were located on LG 5 of the blueberry map and LG 2 of the cranberry map, respectively, suggesting that LG 5 of blueberry aligns with LG 2 of cranberry. Another marker ctg600 mapped to LG 2 of the cranberry map, while the comparable blueberry marker GVC-C600_132 grouped with LG 5 markers. This marker was not placed until the third round and therefore is not shown on the map. This also suggests, however, that LG 2 of the cranberry map is comparable to LG 5 of the blueberry map. Another pair of shared markers, GVC-V22a02_171 and GVC-V22a02_173, and 2ms2a02, was located on LG 7 of the blueberry map and LG 5 of the cranberry map, respectively, suggesting these LG correspond. Finally, the pair of shared markers, GVC-V32g09_292 and 3ms2g09, mapped to LG 9 of blueberry and LG 3 of cranberry, respectively, suggesting these also correspond with each other.
Our research to date has focused on identifying QTL for traits associated with climatic adaptation, cold hardiness, and chilling requirement, in the blueberry mapping population. In this study, we identified one QTL for cold hardiness on LG 4 that was significant at the 5 % error rate in 2009 and 2010. The closest markers to the QTL were EST-PCR marker leaf-00248, followed by berc230. The markers appeared to be located 3 and 5 cM from the QTL in 2009, and 0 and 2 cM from the QTL in 2010. This is the same region of the map that was identified previously as having a putative QTL for cold hardiness (LOD >2) using MAPMAKER/QTL (Rowland et al. 2003a). At that time, the RAPD marker OPAR12b was the closest marker to the QTL; EST-PCR markers leaf-00248 and berc230 had not yet been mapped. The identified QTL explained 17.9 % of the phenotypic variance of the population. In the present study, in 2009 and 2010, the QTL explained 32.9 and 20.1 % of the phenotypic variance, respectively. From our analyses, broad-sense heritability (H) of cold hardiness was high (0.88) in the blueberry mapping population. By dividing the percent of the phenotypic variance explained by H (De Vicente and Tanksley 1993; Lewers et al. 1999), it was determined that the cold hardiness QTL explained 37.4 and 22.9 % of the genotypic variance in 2009 and 2010, respectively (Table 3).
Contigs leaf-00248 and berc230 (from which the EST-PCR markers were derived) have been blasted against the all assembly of the blueberry 454 transcriptome database. Contig leaf-00248 had high similarity to contig all-12494 encoding a predicted peptidyl-prolyl cis–trans isomerase-like protein or cyclophilin with a CP12 (chloroplast protein 12) domain. Berc230 had high similarity to contig all-01955 encoding a hypothetical protein in the alpha/beta hydrolase superfamily. The leaf-00248 and berc230 markers were amplified in Fla4B and absent in W85-20, and presence of the markers was associated with cold sensitivity as would be expected considering the cold hardiness of the parent plants.
Mapping QTL for cold hardiness has been accomplished in several plant species including various cereals (Galiba et al. 1995; Francia et al. 2004; Fisk et al. 2013), legumes (Avia et al. 2013; Tayeh et al. 2013), forestry species (Howe et al. 2000, 2003; Jermstad et al. 2001, 2003; Tsarouhas et al. 2004; Wheeler et al. 2005), and citrus (Weber et al. 2003). Association mapping of cold hardiness has been carried out as well in several species including Douglas fir (Eckert et al. 2009a, b) and Sitka spruce (Holliday et al. 2010). In a few cases, the genes underlying cold hardiness QTL have been identified. For example, variation in the copy number of the CBF gene (Stockinger et al. 1997; Vogel et al. 2005) at Frost Resistance-2 has been found to be associated with levels of freezing tolerance in temperate-climate cereals (Francia et al. 2007; Knox et al. 2010). C-repeat binding factors or CBFs are transcription factors that regulate one, but not the only, cold acclimation pathway in plants (Stockinger et al. 1997; Fowler and Thomashow 2002; Vogel et al. 2005). The association mapping studies in Douglas fir and Sitka spruce found most notable associations of cold hardiness with genes involved in light signal transduction, in the case of autumn cold hardiness in Sitka spruce (Holliday et al. 2010), as well as genes involved in lignin biosynthesis, cell membrane and cell wall architecture, and chaperone function in Douglas fir (Eckert et al. 2009a, b).
To test whether the CBF gene previously isolated from blueberry flower buds during cold acclimation (Naik et al. 2007; Polashock et al. 2010) could be responsible for the cold hardiness QTL identified in our mapping population on LG 4, the gene sequence was blasted against the current blueberry whole genome assembly (Allan Brown, unpublished). The gene was located to one of the scaffolds, and SSR primers were designed to flank SSRs near the gene. The amplified SSR markers, KAN-0009C_160 and KAN-0009C_165, mapped to LG 2 (Fig. 2), however, suggesting that the CBF gene is not responsible for the identified QTL.
In our study, we also successfully identified QTL for chilling requirement. We found one QTL on LG 6 that coincided with the RAPD marker OPP-09b and was significant at the 5 % error rate in 2012 and 2013. A second QTL for chilling requirement was identified only in 2013 located 2 cM from RAPD marker OPP-14d on LG 8, which was significant at the 10 % error rate. Broad-sense heritability of chilling requirement was also high (0.86), like that of cold hardiness. In 2012 and 2013, the QTL at OPP-09b explained 18.6 and 16.7 % of the phenotypic variance, and 21.6 and 19.4 % of the genotypic variance, respectively. In 2013, the second QTL near OPP-14d explained 14.6 % of the phenotypic variance and 16.9 % of the genotypic variance.
Also, within the chilling requirement QTL, confidence region on LG 6 was the RAPD marker OPB-05a (9 cM from the QTL) and the EST-PCR marker berc826 (13 cM from the QTL). Contig berc826 was blasted against the all assembly of the blueberry 454 transcriptome database and found to have high similarity to contig all-02175, which encodes a putative proline-rich cell wall protein.
The marker OPP-09b, near the strongest QTL for chilling requirement on LG 6, was amplified in Fla4B, absent in W85-20, and associated with lower chilling requirement. The markers further from the QTL, OPB-05a, and berc826, were amplified in W85-20 and associated with higher chilling requirement. The marker OPP-14d, closest to the weaker QTL on LG 8 (detected only in 2013), was amplified in W85-20, but surprisingly was associated with lower chilling requirement. This suggests that the parent W85-20 is heterozygous for at least one gene that contributes to chilling requirement.
Quantitative trait loci for chilling requirement have been identified in a few fruit crops, including peach (Fan et al. 2010), almond (Sanchez-Perez et al. 2012), and cherry (Quero-Garcia et al. 2010). Some of the identified chilling requirement QTL appear to be in common among these Prunus species, such as a significant QTL on LG 4. Probably the best studied, however, is a major QTL identified on LG 1 of peach (Fan et al. 2010), which coincides with the EVG (evergrowing mutation) locus (Wang et al. 2002; Bielenberg et al. 2004, 2008). The genes responsible for this QTL (and mutation) appear to be a group of DAM [dormancy-associated Short Vegetative Phase (SVP)-like MADS-box] genes (Li et al. 2009; Jimenez et al. 2010; Fan et al. 2010). In addition, the expression of DAM4, DAM5, and DAM6 appears to be correlated with bud dormancy release in a group of peach cultivars with varying chilling requirements (Leida et al. 2012). On the other hand, in a recent association mapping study done on 104 peach landrace accessions from China, no association was found between chilling requirement and the region around the QTL identified in Fan et al. (2010) and the EVG locus (Cao et al. 2012).
To test whether DAM-like genes are responsible for the major chilling requirement QTL identified in our mapping population, DAM4, DAM5, and DAM6 gene sequences from peach were blasted against the current blueberry whole genome assembly (Allan Brown, unpublished), and SSR primers were designed to flank SSRs near the genes. Of the SSR markers that amplified, some were not polymorphic and therefore could not be mapped, while others did not map near the chilling requirement QTL (data not shown). Although this work is ongoing, it so far suggests that these DAM genes are not responsible for the identified QTL.
Efforts are currently underway to anchor the genetic linkage map of our population to the assembly of the whole genome sequence of V. corymbosum W85-20 to allow us to directly identify genes in the vicinity of QTL for cold hardiness and chilling requirement. In addition, we are continuing to map other candidate genes for these traits, like the E3 ubiquitin-protein ligase gene family found near a QTL for chilling requirement in apple (Celton et al. 2011). Efforts to map fruit quality traits that are segregating in the blueberry population are also in progress.
Abbreviations
- CH:
-
Cold hardiness
- CR:
-
Chilling requirement
- CR50:
-
Chill units resulting in 50 % floral bud break
- EST-PCR:
-
Expressed sequence tag-polymerase chain reaction
- H :
-
Broad-sense heritability
- HRM:
-
High-resolution melting
- IM:
-
Interval mapping
- LOD:
-
Logarithm of the odds
- LT50:
-
Lethal temperature causing 50 % injury
- PM:
-
Permutation test
- QTL:
-
Quantitative trait locus/loci
- RAPD:
-
Randomly amplified polymorphic DNA
- rMQM:
-
Restricted multiple QTL mapping
- SNP:
-
Single nucleotide polymorphism
- SRA:
-
Sequence read archive
- SSR:
-
Simple sequence repeat
References
Arora R, Rowland LJ, Lehman JS, Lim CC, Panta GR, Vorsa N (2000) Genetic analysis of freezing tolerance in blueberry (Vaccinium section Cyanococcus). Theor Appl Genet 100:690–696
Arora R, Rowland LJ, Ogden EL, Dhanaraj AL, Marian CO, Ehlenfeldt MK, Vinyard B (2004) Dehardening kinetics, bud development, and dehydrin metabolism in blueberry (Vaccinium spp.) cultivars during deacclimation at constant, warm temperatures. J Am Soc Hortic Sci 129:667–674
Avia K, Pilet-Nayel M-L, Bahrman N, Baranger A, Delbreil B, Fontaine V, Hamon C, Hanocq E, Niarquin M, Sellier H, Vuylsteker C, Prosperi J-M, Lejeune-Henaut I (2013) Genetic variability and QTL mapping of freezing tolerance and related traits in Medicago truncatula. Theor Appl Genet 126:2353–2366
Basu A, Du M, Leyva MJ, Sanchez K, Betts NM, Wu M, Aston CE, Lyons TJ (2010) Blueberries decrease cardiovascular risk factors in obese men and women with metabolic syndrome. J Nutr 140:1582–1587
Bielenberg DG, Wang Y, Fan S, Reighard GL, Scorza R, Abbott AG (2004) A deletion affecting several gene candidates is present in the evergrowing peach mutant. J Hered 95:436–444
Bielenberg DG, Wang Y, Li Z, Zhebentyayeva T, Fan S, Reighard GL, Scorza R, Abbott AG (2008) Sequencing and annotation of the evergrowing locus in peach [Prunus persica (L.) Batsch] reveals a cluster of six MADS-box transcription factors as candidate genes for regulation of terminal bud formation. Tree Genet Genomes 4:495–507
Boches P, Bassil NV, Rowland LJ (2006) Genetic diversity in the highbush blueberry Vaccinium corymbosum L. evaluated with microsatellite markers. J Am Soc Hortic Sci 131:674–686
Bushakra JM, Stephens MJ, Atmadjaja AN, Lewers KS, Symonds VV, Udall JA, Chagne D, Buck EJ, Gardiner SE (2012) Construction of black (Rubus occidentalis) and red (R. idaeus) raspberry linkage maps and their comparison to the genomes of strawberry, apple, and peach. Theor Appl Genet 125:311–327
Cao K, Wang L, Zhu G, Fang W, Chen C, Luo J (2012) Genetic diversity, linkage disequilibrium, and association mapping analyses of peach (Prunus persica) landraces in China. Tree Genet Genomes 8:975–990
Celton JM, Martinez S, Jammes MJ, Bechti A, Salvi S, Legave JM, Costes E (2011) Deciphering the genetic determinism of bud phenology in apple progenies: a new insight into chilling and heat requirement effects on flowering dates and positional candidate genes. N Phytol 192:378–392
Chagne D, Gasic K, Crowhurst RN, Han Y, Bassett HC, Bowatte DR, Lawrence TJ, Rikkerink EHA, Gardiner SE, Korban SS (2008) Development of a set of SNP markers present in expressed genes of the apple. Genomics 92:353–358
Chakravarti A, Lasher LK, Reefer JE (1991) A maximum likelihood method for estimating genome length using genetic linkage data. Genetics 128:175–182
Cho E, Seddon JM, Rosner B, Willett WC, Hankinson SE (2004) Prospective study of intake of fruits, vegetables, vitamins and carotenoids and risk of age-related maculopathy. Arch Ophthalmol 122:883–892
De Vicente MC, Tanksley S (1993) QTL analysis of transgressive segregation in an interspecific tomato cross. Genetics 134:585–596
Dhanaraj AL, Slovin JP, Rowland LJ (2004) Analysis of gene expression associated with cold acclimation in blueberry floral buds using expressed sequence tags. Plant Sci 166:863–872
Dhanaraj AL, Alkharouf NW, Beard HS, Chouikha IB, Matthews BF, Wei H, Arora R, Rowland LJ (2007) Major differences observed in transcript profiles of blueberry during cold acclimation under field and cold room conditions. Planta 225:735–751
Eckert AJ, Bower AD, Wegrzyn JL, Pande B, Jermstad KD, Krutovsky KV, Clair JBS, Neale DB (2009a) Association genetics of coastal Douglas fir (Pseudotsuga menziesu var. menziesii, Pinaceae). I. Cold-hardiness related traits. Genetics 182:1289–1302
Eckert AJ, Wegrzyn JL, Pande B, Jermstad KD, Lee JM, Liechty JD, Tearse BR, Krutovsky KV, Neale DB (2009b) Multilocus patterns of nucleotide diversity and divergence reveal positive selection at candidate genes related to cold hardiness in coastal Douglas fir (Pseudotsuga menziesu var. menziesii). Genetics 183:289–298
Fan S, Bielenberg DG, Zhebentyayeva TN, Reighard GL, Okie WR, Holland D, Abbott AG (2010) Mapping quantitative trait loci associated with chilling requirement, heat requirement and bloom date in peach (Prunus persica). N Phytol 185:917–930
Fear CD, Lauer FI, Luby JJ, Stucker RL, Stushnoff C (1985) Genetic components of variance for winter injury, fall growth cessation, and off-season flowering in blueberry progenies. J Am Soc Hortic Sci 110:262–266
Fisk SP, Cuesta-Marcos A, Cistue L, Russell J, Smith KP, Baenziger S, Bedo Z, Corey A, Filichkin T, Karsai I, Waugh R, Hayes PM (2013) FR-H3: a new QTL to assist in the development of fall-sown barley with superior low temperature tolerance. Theor Appl Genet 126:335–347
Flinn CL, Ashworth EN (1994) Blueberry flower hardiness is not estimated by differential thermal analysis. J Am Soc Hortic Sci 119:295–298
Fowler S, Thomashow MF (2002) Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell 14:1675–1690
Francia E, Rizza F, Cattivelli L, Stanca AM, Galiba G, Toth B, Hayes PM, Skinner JS, Pecchioni N (2004) Two loci on chromosome 5H determine low-temperature tolerance in a ‘Nure’ (winter) × ‘Tremois’ (spring) barley map. Theor Appl Genet 108:670–680
Francia E, Barabaschi D, Tondelli A, Laido G, Rizza F, Stanca AM, Busconi M, Fogher C, Stockinger EJ, Pecchioni N (2007) Fine mapping of a HvCBF gene cluster at the frost resistance locus Fr-H2 in barley. Theor Appl Genet 115:1083–1091
Galiba G, Quarrie SA, Sutka J, Morgounov A, Snape JW (1995) RFLP mapping of vernalization (Vrn1) and frost resistance (Fr1) on chromosome 5A of wheat. Theor Appl Genet 90:1174–1179
Galletta GJ, Ballington JR (1996) Blueberries, cranberries and lingonberries. In: Janick J, Moore JN (eds) Fruit breeding. Vine and small fruit crops, vol II. Wiley, New York, pp 1–107
Georgi L, Johnson-Cicalese J, Honig J, Das SP, Rajah VD, Bhattacharya D, Bassil N, Rowland LJ, Polashock J, Vorsa N (2013) The first genetic map of the American cranberry: exploration of synteny conservation and quantitative trait loci. Theor Appl Genet 126:673–692
Holland JB, Nyquist WE, Cervantes-Martinez CT (2003) Estimating and interpreting heritability for plant breeding: an update. Plant Breed Rev 22:9–112
Holliday JA, Ritland K, Aitken SN (2010) Widespread, ecologically relevant genetic markers developed from association mapping of climate-related traits in Sitka spruce (Picea sitchensis). N Phytol 188:501–514
Howe GT, Saruul P, Davis J, Chen THH (2000) Quantitative genetics of bud phenology, frost damage, and winter survival in an F2 family of hybrid poplars. Theor Appl Genet 101:632–642
Howe GT, Aitken SN, Neale DB, Jermstad KD, Wheeler NC, Chen THH (2003) From genotype to phenotype: unraveling the complexities of cold adaptation in forest trees. Can J Bot 81:1247–1266
Jaakola L, Maatta K, Pirttila AM, Torronen R, Karenlampi S, Hohtola A (2002) Expression of genes involved in anthocyanin biosynthesis in relation to anthocyanin, proanthocyanidin, and flavonol levels during bilberry fruit development. Plant Phys 130:729–739
Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF (1998) Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280:104–106
Jermstad KD, Bassoni DL, Wheeler NC, Anekonda TS, Aitken SN, Adams WT, Neale DB (2001) Mapping of quantitative loci controlling adaptive traits in coastal Douglas-fir. II. Spring and fall cold-hardiness. Theor Appl Genet 102:1152–1158
Jermstad KD, Bassoni DL, Jech KS, Ritchie GA, Wheeler NC, Neale DB (2003) Mapping of quantitative loci controlling adaptive traits in coastal Douglas Fir. III. Quantitative trait loci-by-environment interactions. Genetics 165:1489–1506
Jimenez S, Li Z, Reighard GL, Bielenberg DG (2010) Identification of genes associated with growth cessation and bud dormancy entrance using a dormancy-incapable tree mutant. BMC Plant Biol 10:25
Kalt W, Joseph JA, Shukitt-Hale B (2007) Blueberries and human health. A review of the current research. J Am Pomol Soc 61:151–160
Knapp SJ, Stroup WW, Ross WM (1985) Exact confidence intervals for heritability on a progeny mean basis. Crop Sci 25:192–194
Knox AK, Dhillon T, Cheng H, Tondelli A, Pecchioni N, Stockinger EJ (2010) CBF gene copy number variation at Frost Resistance-2 is associated with levels of freezing tolerance in temperate-climate cereals. Theor Appl Genet 121:21–35
Krebs SL, Hancock JF (1988) The consequences of inbreeding on fertility in Vaccinium corymbosum L. J Am Soc Hortic Sci 113:914–918
Leida C, Romeu JF, Garcia-Brunton J, Rios G, Badenes ML (2012) Gene expression analysis of chilling requirements for flower bud break in peach. Plant Breed 131:329–334
Lewers KS, Crane EH, Bronson CR, Schupp JM, Keim P, Shoemaker RC (1999) Detection of linked QTL for soybean brown stem rot resistance in ‘BSR 101’ as expressed in a growth chamber environment. Mol Breed 5:33–42
Li Z, Reighard GL, Abbott AG, Bielenberg DG (2009) Dormancy-associated MADS genes from the EVG locus of peach [Prunus persica (L.) Batsch] have distinct seasonal and photoperiodic expression patterns. J Exp Bot 60:3521–3530
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10:1391–1406
Manly BFJ (1997) Randomization, bootstrap and Monte Carlo methods in biology, 2nd edn. Chapman & Hall, London
Montgomery J, Wittwer CT, Palais R, Zhou L (2007) Simultaneous mutation scanning and genotyping by high-resolution DNA melting analysis. Nat Protoc 2:59–66
Naik D, Dhanaraj AL, Arora R, Rowland LJ (2007) Identification of genes associated with cold acclimation in blueberry (Vaccinium corymbosum L.) using a subtractive hybridization approach. Plant Sci 173:213–222
Polashock JJ, Arora R, Peng Y, Naik D, Rowland LJ (2010) Functional identification of a C-repeat binding factor transcriptional activator from blueberry associated with cold acclimation and freezing tolerance. J Am Soc Hortic Sci 135:40–48
Prior RL, Cao GH, Martin A, Sofic E, McEwen J, O’Brien C, Lischner N, Ehlenfeldt M, Kalt W, Krewer G, Mainland CM (1998) Antioxidant capacity as influenced by total phenolic and anthocyanin content, maturity, and variety of Vaccinium species. J Agric Food Chem 46:2686–2693
Qu L, Hancock JF (1997) Randomly amplified polymorphic DNA- (RAPD-) based genetic linkage map of blueberry derived from an interspecific cross between diploid Vaccinium darrowii and tetraploid V. corymbosum. J Am Soc Hortic Sci 122:69–73
Quero-Garcia J, Le Dantec L, Fodor A, Reignier A, Capdevilla G, Joly J, Tauzin Y, Fouilhaux L, Dirlewanger E (2010) QTL detection for fruit quality and phenological characters in sweet cherry. In: 5th international rosaceae genomics conference, Stellenbosch, South Africa
Reed GH, Wittwer CT (2004) Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin Chem 50:1748–1754
Rowland LJ, Levi A (1994) RAPD-based genetic linkage map of blueberry derived from a cross between diploid species (Vaccinium darrowi × V. elliottii). Theor Appl Genet 87:863–868
Rowland LJ, Lehman JS, Levi A, Ogden EL, Panta GR (1998) Genetic control of chilling requirement in blueberry. In: Cline WO, Ballington JR (eds) Proceedings of the 8th North American Blueberry Research and extension worker’s conference, North Carolina State University, Raleigh, NC, USA, pp 258–267
Rowland LJ, Ogden EL, Arora R, Lim C-C, Lehman JS, Levi A, Panta GR (1999) Use of blueberry to study genetic control of chilling requirement and cold hardiness in woody perennials. HortScience 34:1185–1191
Rowland LJ, Mehra S, Dhanaraj A, Ogden EL, Arora R (2003a) Identification of molecular markers associated with cold tolerance in blueberry. Acta Hortic 625:59–69
Rowland LJ, Mehra S, Dhanaraj AL, Ogden EL, Slovin JP, Ehlenfeldt MK (2003b) Development of EST-PCR markers for DNA fingerprinting and genetic relationship studies in blueberry (Vaccinium, section Cyanococcus). J Am Soc Hortic Sci 128:682–690
Rowland LJ, Ogden EL, Ehlenfeldt MK, Vinyard B (2005) Cold hardiness, deacclimation kinetics, and bud development among 12 diverse blueberry genotypes under field conditions. J Am Soc Hortic Sci 130:508–514
Rowland LJ, Hancock JF, Bassil NV (2011) Blueberry. In: Folta K, Kole C (eds) Genetics, genomics and breeding of berries. Science, Enfield, pp 1–40
Rowland LJ, Alkharouf N, Darwish O, Ogden EL, Polashock JJ, Bassil NV, Main D (2012) Generation and analysis of blueberry transcriptome sequences from leaves, developing fruit, and flower buds from cold acclimation through deacclimation. BMC Plant Biol 12:46. doi:10.1186/1471-2229-12-46
Sanchez-Perez R, Dicenta F, Martinez-Gomez P (2012) Inheritance of chilling and heat requirements for flowering in almond and QTL analysis. Tree Genet Genomes 8:379–389
Schuelke M (2000) An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol 18:233–234
Spiers JM (1978) Effect of stage of bud development on cold injury in rabbiteye blueberry. J Am Soc Hortic Sci 103:452–455
Stockinger EJ, Gilmour SJ, Thomashow MF (1997) Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA 94:1035–1040
Tayeh N, Bahrman N, Devaux R, Bluteau A, Prosperi J-M, Delbreil B, Lejeune-Henaut I (2013) A high-density genetic map of the Medicago truncatula major freezing tolerance QTL on chromosome 6 reveals colinearity with a QTL related to freezing damage on Pisum sativum linkage group VI. Mol Breed 32:279–289
Tsarouhas V, Gullberg U, Lagercrantz U (2004) Mapping of quantitative trait loci (QTLs) affecting autumn freezing resistance and phenology in Salix. Theor Appl Genet 108:1335–1342
Van Ooijen JW (2004) MapQTL® 5: software for the mapping of quantitative trait loci in experimental populations of diploid species. Kyazma B.V., Wageningen
Van Ooijen JW (2006) JoinMap® 4: software for the calculation of genetic linkage maps in experimental populations. Kyazma B.V., Wageningen
Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF (2005) Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J 41:195–211
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wang Y, Georgi LL, Reighard GL, Scorza R, Abbott AG (2002) Genetic mapping of the evergrowing gene in peach [Prunus persica (L.) Batsch]. J Hered 93:352–358
Weber CA, Moore G, Deng Z, Gmitter FG Jr (2003) Mapping freeze tolerance quantitative trait loci in a Citrus grandis × Poncirus trifoliata F1 pseudo-testcross using molecular markers. J Am Soc Hortic Sci 128:508–514
Wheeler NC, Jermstad KD, Krutovsky K, Aitken SN, Howe GT, Krakowski J, Neale DB (2005) Mapping of quantitative trait loci controlling adaptive traits in coastal Douglas-fir. IV. Cold-hardiness QTL verification and candidate gene mapping. Mol Breed 15:145–156
Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ (2003) High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem 49:853–860
Acknowledgments
The authors gratefully acknowledge all the high school and college students who participated in this project as part of an internship or other training opportunity. We specifically would like to acknowledge the students from the University of Maryland (Brianna Driscoll, Dana Robinson, and Jenny Lindvall), who worked in Dr. Rowland’s lab on the mapping project, and the students from Davidson College (Mark Angel, Erich Baker, Spencer Chadinha, Stewart Dalton, Aaron Deal, Catherine Doyle, Tim Keating, David Lloyd, Austin Mudd, Mike Nuttle, Shamita Punjabi, and Daniel Tuerff), who worked in Dr. Campbell’s class to design SSRs near genes of interest. We would also like to thank Barbara Gilmore, April Nyberg, Elisabeth Alperin, and Jeremy Jones, who were involved in screening the mapping parents for SSR polymorphism. This project was partially funded by USDA-ARS Project 1245-21000-185-00D and USDA-CSREES Specialty Crop Research Initiative Grant 2008-51180-04861 entitled ‘Generating Genomic Tools for Blueberry Improvement.’ Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture or any of the other agencies involved in this research.
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rowland, L.J., Ogden, E.L., Bassil, N. et al. Construction of a genetic linkage map of an interspecific diploid blueberry population and identification of QTL for chilling requirement and cold hardiness. Mol Breeding 34, 2033–2048 (2014). https://doi.org/10.1007/s11032-014-0161-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-014-0161-9