Abstract
α-Amylase plays a key role in seed germination. Activity of α-amylase determines levels of starch degradation, seed germination, and pre-harvest sprouting (PHS), which is a serious problem in wheat production. In this study, we isolated and characterized high PI amylase coding genes from the wheat cultivar Chuanmai32 (PHS susceptible) and the synthetic wheat SHW-L1 (PHS resistant). amy1 gene sequences were 1,459 a long and contained three exons and two introns. Phylogenetic analysis revealed that homologous genes obtained from the two accessions were extremely conserved and belonged to barley AMY2-1 subgroup. Six nucleotide substitutions were detected in the exon regions between the two amy1 genes. The amino acid substitutions Lys364/Arg364 and Arg366/Trp366 occurred in the C-terminal region, which is present in the anti β-sheet three-dimensional structure of AMY1. Expression profiling of amy1 indicated that mRNA transcript accumulation began at a late stage of grain development. amy1 transcript accumulation in Chuanmai32 was 4.32- and 18.36-fold higher than observed in SHW-L1 at DPA25 and DPA30, respectively. Two significant expression quantitative trait loci (eQTLs) on chromosome 1BS and one on 3DS were characterized by expression analysis of amy1 transcripts and genetic analysis of SHW-L1/Chuanmai32-derived recombinant inbred lines. The genes that encoded high PI amylase were located near the centromere on chromosomes 6AL/6BL/6DL. These results suggest that these eQTL regions may provide candidate genes that play potential roles in regulating PHS through effects on amy1 expression.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pre-harvest sprouting (PHS) is defined as the germination of grains in the spike before harvest in wet conditions and is one of the main causes of damage in wheat (Triticum spp.) production areas. PHS severely reduces the yield and quality of grains (Buchanan and Nicholas 1980; Varughese et al. 1986; Derera 1989). α-Amylase (EC 3.2.1.1) plays a key role in plant metabolism though the hydrolyzation of starch. Its function is to convert the 1, 4-α endoglycolytic of amylose and amylopectin into glucose, maltose, and small dextrins and oligosaccharides. Cereals contain many isozymic forms of α-amylase, these are classified into two main groups based on isoelectric point. AMY2 is classified as a low pI α-amylase group and AMY1 as a high pI α-amylase group (Huang et al. 1992; Huttly et al. 1988; Lazarus et al. 1985). Amy1 gene was located on 6A/B/D in wheat, while its encoded enzymes were called AMY2 among the most described α-amylases in barley. They are known to play essential roles in the germination and malting process through hydrolyzation of storage starch granules present in the endosperm (Knox et al. 1987; Gale et al. 1983; Cheng et al. 2013).
α-Amylase activity differs during grain development: peaking at around DPA15 (days post anthesis) and then decreasing (Mares and Oettler 1991). In wheat, both AMY1 and AMY2 are detected in germinating grains, but the isozymes are active at distinct stages: AMY1 isozymes are more active at the beginning of germination (1–2 days), while AMY2 isozymes increase later (third day) (Sargeant 1980). amy1, amy2, and amy3 gene transcripts accumulate at different periods in the grains (Kruger 1976; Baulcombe et al. 1987). The transcripts of amy2 are detected in the pericarp of developing grains (Sargeant 1980; Gale and Ainsworth 1984; Daussant and Renard 1987; Lazarus et al. 1985). Mares and Oettler (1991) found that different genotypes have different activities in the grain at about DPA30; this indicates that the expression of amy1 at DPA30 is associated with the different physiological activities of grain and may affect the potential germination ability of grain (Mares and Oettler 1991).
Characterization of AMY2 s and their coding gene sequences in wheat will help further the understanding of key genes underlying PHS. In this study, we cloned amy1 from the cultivar Chuanmai32 (PHS susceptible) and the synthetic wheat SHW-L1 (PHS resistant). We describe the expression pattern of amy1 at different grain developmental stages and identify their potential relationship with PHS resistance. Furthermore, QTL expression analysis was performed to detect regions regulating α-amylase gene expression in recombinant inbred lines (RILs) obtained from the synthetic wheat and cultivar.
Materials and methods
Plant material and linkage map structure
PHS-susceptible hexaploid wheat cultivar Chuanmai32 and medium PHS-resistant synthetic hexaploid wheat SHW-L1 (synthesized from Triticum turgidum AS2255 and Aegilops tauschii AS60) were used for amy2 sequence characterization and expression level analysis. RILs derived from these two accessions were used to characterize regions that regulated amy1 expression levels. The linkage map of the RILs was constructed in a previous study and contains 1,794 diversity arrays technology (DArT) markers and 68 simple sequence repeat (SSR) markers (Yu and Chen 2013). These markers covered all 21 chromosomes at an average of 88.7 loci/chromosome. There were 961 loci (1.27 cM/marker) on sub-genome D, 539 (2.49 cM/marker) on sub-genome B, and 362 (3.33 cM/marker) on sub-genome A.
Cloning and phylogenetic analysis of the amy1 gene
Nucleotide sequences from Barley amy genes were used as reference sequences to search for homologous genes in the wheat whole-genome shotgun sequence (WGS) database using the Basic Local Alignment Search Tool (http://blast.ncbi.nlm.nih.gov/). Primers (Invitrogen, Shanghai, China) were designed for PCR amplification of the amy1 genomic sequence. PCR was performed with the following conditions: initial denaturing at 95 °C for 5 min, followed by thirty-five cycles of denaturing at 95 °C for 45 s, annealing at 60 °C for 45 s, extension at 72 °C for 1 min, and a final extension at 72 °C for 7 min. The forward primer (Pf) contained nucleotide sequences following the start codon ATG, and the reverse primer (Pr) was eight nucleotides before the stop TAA codon (Table 1). The desired amplified fragment covered most of the amy1 gene sequence, including all introns and exons.
As there were no previously available phylogenetic studies of α-amylase amino acid sequences obtained from wheat, sequences were analyzed using the Kimura 2-parameter model included in MEGA 4.0 (http://www.megasoftware.net) to identify possible phylogenetic clades.
Expression assay and eQTL analysis of amy1 genes in RILs
Common phenological traits in crops are often controlled by genetic loci known as quantitative trait loci (QTLs). Similarly, mRNA transcripts have been used as traits for expression quantitative trait loci (eQTL) analysis by measuring transcript expression levels across genes in cereal populations such as wheat, barley, and rice (Jansen and Nap 2001; Munkvold et al. 2013; Jordan et al. 2007; Chen et al. 2010; Potokina et al. 2008; Wang et al. 2010; Vergne et al. 2010). For our expression assays, a pair of primers in the relevant exon regions of amy1 was used to amplify a 148 bp sequence. Expression profiles of amy1 were measured from the two parents and randomly selected lines at five stages of seed development (5, 15, 20, 25, and 30 DPA). Immature seeds were collected from the center of the spikes and rapidly put into liquid nitrogen and then stored at −70 °C. Total RNA was extracted from each sample using an RNA extraction kit (Takara, Dalian, China) according to the manufacturer’s instructions. First-strand cDNA was synthesized using a PrimeScript™ 1st Strand cDNA Synthesis Kit (Takara). cDNA sampling was performed in duplicate, and SYBR Green (TIANGEN, Beijing, China) used for real-time PCR (MyiQ Real-Time PCR Detection System, Bio-Rad, USA).
Six ten-proportion dilutions of recombination plasmid templates were used to make a standard curve to calculate amplification efficiency (E) and to control amplifications. For all samples, three reference genes: TaActin, Ta.14126.1, and Ta.7894.3.A1_at were used as internal control genes (Long et al. 2010). Data were analyzed using Bio-Rad iQ5 Software 2.0 (Bio-Rad). The level of each template was measured against the internal control genes and the geometric mean statistically calculated. The relative expression quantity of each sample was calculated using the E −ΔΔCtmethod (Pfaffl 2001). The relative expression quantities of the RILs were used for eQTL analysis.
Significance of the eQTLs was calculated by WinQTL Cart 2.5 software (North Carolina State University, Raleigh, NC, USA) using the composite interval mapping (CIM) method (Wang et al. 2007). The suitable scan speed was 2 cM. Analysis was implemented using WinQTL Cart 2.5 by setting the control parameter to model 6 (standard model), forward regression, 10 cM window, and five makers as control. The threshold was automatically calculated by the software based on 1,000 permutation times to avoid invalid loci or loss of affective loci at the significance threshold of p < 0.05. Final permutation times of eQTL detection were 500, p < 0.05.
Results
Characterization of amy1 from pre-harvest sprouting resistant and susceptible wheat
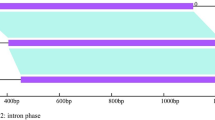
Gene-specific primers were used to amplify amy1 gene (KJ470677 and KJ470678) sequences from the genomic DNA of PHS-susceptible cultivar Chanmai32 and PHS-resistant synthetic wheat SHW-L. The isolated nucleotide sequences of amy1 were both 1,459 bp, with open reading frames (ORFs) of 423 amino acid residues. Both contained three exons and two introns (Fig. 1).
The two sequences shared 99.59 % identity and had six single-nucleotide polymorphisms (SNPs), of which four were located in the second exon and two on the third (Fig. 1). No frameshift mutations were found in the exon regions, indicating that the ORF could completely translate the amino acid sequence. The positions of each SNP in the sequence, whether synonymous (silent) or nonsynonymous (replacement), were further determined. Two amino acid residues variations between two genotypes were detected: Lys364/Arg364 and Arg366/Trp366, the other four SNPs did not change the amino acid primary structure (Fig. 2).
The α-amylases translated by the cloned amy1 genes were compared with known barley and wheat α-amylases: X15226.1, X15227.1, M17125.1, M17126.1, J04202.1, J01236.1, FN179390.1, FN179391.1, FN179392.1, K02637.1, Triticum urartu G1812_contig 778314, T. urartu G1812_contig 953858, and Aegilops tauschii AL8/78_contig 35149. Phylogenetic analysis of these proteins was then performed using MEGA 4.0 software (Tamura et al. 2007). Four groups were found to be clustered based on the neighbor-joining method; these were referenced as AMY1, AMY2, AMY3, and AMY4 groups (Fig. 3). The AMY2 group was divided into two subgroups: AMY2-1 and AMY2-2. Translated sequences of cloned wheat amy1 sequences in this study were placed into the barley AMY2-1 subgroup and shared nearest evolution distances with T.urartu_contig 953858, T.urartu_contig 778314, and four barley α-amylases.
Phylogeny analyses of AMY1 amino acid sequences isolated from SHW-L1 and Chuanmai32 with other α-amylase genes from wheat and barley. Sequences were derived from the NCBI nucleotide database and whole-genome shotgun database, and analyses performed by MEGA4.0 software. All sequences are shown with associated species name and GenBank accessions
A conserved domain search of the NCBI website (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) indicated that the active site of α-amylase was located from the 26th to the 376th aa, a span of 350 aa. The comparison revealed that AMY1, AMY2, and AMY3 were conserved in this domain, but AMY4 was not. One short domain (GYAYILTHPG) was completely conserved in all 17 sequences and flanking amino acids only showed small variations (Fig. 2).
Expression pattern of amy1 in pre-harvest sprouting resistant and susceptible wheat genotypes
The expression levels of amy1 at five time points (5, 15, 20, 25, and 30 DPA) during grain development were examined by RT-PCR. The expression patterns of amy1 in the two genotypes were different (Fig. 4). In PHS-resistant wheat SHW-L1, peak amy1 transcript appeared at DPA20. This was consistent with previous studies that reported the peak of amylase activity occurred at DPA15–DPA23 during grain development (Huttly et al. 1988; Cheng et al. 2013). There was no accumulation of amy1 transcript in Chuanmai32 in the first 20 days; this was followed by a rapid accumulation at DPA25 and a peak at DPA30. The expression of amy1 was higher in Chuanmai32 at DPA25 (4.3-fold) and DPA30 (18.4-fold) compared with SHW-L1.
Identification of regions controlling amy1 mRNA accumulation
Genomic regions controlling the expression of amy1 genes were characterized at times that showed significance expression differences between the SHW-L1 and Chuanmai32 parents. Expression data at DPA20, DPA25, and DPA30 were used for eQTL analysis. Two eQTL regions located near the centromere on chromosome 1B were detected at DPA25 and DPA30; these were designated as eqamy1-DPA25.1B and eqamy1-DPA30.1B. Although the eQTLs detected at these two stages had similar genetic distances (approximately 7 cM) they could be distinguished by different makers (gpw7422/barc181 at DPA25; wPt-741274 at DPA30). Another eQTL was detected near the centromere of chromosome 3DS at DPA30, this was designated as eqamy1-DPA30.3D. This eQTL was located at the flanking markers wPt-8914 and wPt-0327 (Fig. 5). The identified eQTLs had negative additive effects, indicating that eQTLs that could decrease expression of amy1 were derived from synthetic wheat SHW-L1.
Discussion
Several high pI α-amylase and its corresponding genes have been characterized in barley (Rodenburg et al. 1994). However, there is limited information available for high PI α-amylase nucleotide sequences in wheat. Because both wheat and barley belong to the Triticeae tribe, we proposed to isolate amy1 from wheat based on barley homology. Two amy1 gene sequences from two accessions with different PHS resistance, a Sichuan cultivar wheat and a synthetic wheat, were obtained using specific primers designed against the wheat whole-genome shotgun database. Following phylogenetic analysis of 17 α-amylase amino acid sequences derived from the NCBI database, AMY1, AMY2, AMY3, and AMY4 could be separated into four groups. In addition, two subgroups were separated from the AMY2 group. The two newly identified high PI α-amylase sequences belonged to the AMY2-1 subgroup of the barley AMY2 family recognized as amy1 in wheat.
A potential way to obtain information about functional constraints is via evolutionary comparisons; it is generally believed that conserved residues are more likely to be functionally significant than nonconserved residues (Garg et al. 1999). All six SNPs detected in the two new amy1 sequences were located in exon regions, while intron regions of the two sequences showed high similarity. The possibility of alternative splicing in transcription is very low (Fedorova and Fedorov 2003). The SNPs in the α-amylase genes resulted in two amino acid variations; both were located at the C-terminal of the protein, in the onset of the β-sheet region (Kadziola et al. 1994). These variations are likely to be the cause of functional differences between the two genotypes, as the other domains were conserved.
Developmentally, degradation of starch into soluble sugar by amylase promotes grain transition to germination. Therefore, amylase quantity is associated with PHS damage (Masojć and Milczarski 2009). Both amy1 and amy2 are expressed in the developing grain (Colin M. Lazarus et al. 1985; Huttly et al. 1988), and it has been recently reported that there is a relationship between amy1 expression and germination characteristics at the after-ripening stage (De Laethauwer et al. 2013). There were no transcripts detected in the two tested genotypes in the first 15 days, this is mainly because of the use of accumulated reserves during this period. At the end of the grain development stage, amy1 transcripts accumulated at a larger quantity in Chuanmai32 than in SHW-L1. This indicated that starch hydrolyzed by high PI amylase in the PHS-susceptible genotype was quicker than in PHS-resistance genotypes, and especially at DPA30. This phenomenon suggests that PHS-susceptible genotypes germinate easier under suitable conditions. Moreover, this difference could prove a candidate feature for selection of PHS-resistant varieties. Other cereals exhibit similar patterns of amy1 isoform expression in grain development stages to the present study. For example, no AMY2 isoform could be detected until 24 DPA in barley (MacGregor and Dushnicky 1989). We found that amy1 expressed at very low levels in wheat, and this is consistent with data obtained from other wheat genotypes (Cheng et al. 2013).
Late maturity a-amylase (LMA) is a genetic defect that can result in low falling number and high levels of a-amylase at middle to late stage of grain development (Mares and Mrva 2008; Barrero et al. 2013). Therefore, high peak at DPA25 and DPA30 of amy1 transcripts in Chuanmai32 were probably caused by LMA, while relatively low expression of amy1 in SHW-L1 was due to low level of LMA.
Generally, α-amylase is induced by GA and suppressed by ABA in wheat and barley (Higgins et al. 1982; Tregear et al. 1995; Huttly et al. 1992; Gómez-Cadenas et al. 2001; Barrero et al. 2013; Appleford et al. 2007). Therefore, low amy1 mRNA accumulation in Chuanmai32 suggests that the ABA signal prevails the GA signal in the SHW-L1 genetic background. We were able to identify regions controlling amy1 expression using eQTL analysis. This genetic method enables quick identification of regions linked to candidate genes that function upstream of amy1 transcript or the real position of amy1 on chromosomes (Jansen and Nap 2001). Many transcriptional factors involved in ABA signaling have been characterized in wheat. Vp1/ABI3 has an important function in improving PHS and is located on the long arm of 3A/3B/3D (Bailey et al. 1999; Yang et al. 2007; Nakamura and Toyama 2001; Osa et al. 2003). Many QTLs associated with the PHS detected on chromosome 3D were consistent with the TaVp1 gene (Munkvold et al. 2009; Fofana et al. 2009; Kulwal et al. 2004). Other homologous genes, such as TaABI1, TaABI5, and TaMFT also mapped to the 3A/3B/3D chromosomes (Nakamura et al. 2007, 2011). Among these genes or QTLs, all genes except TaMFT were located on the telomere of 3DL. In our study, an eQTL locus was detected near the centromere of chromosome 3DL. Comparative genetic analysis with barley suggests that TaABI1 is the closest homologous gene to the centromere of 3DL. This gene encodes a phosphatase 2C protein, a negative regulator of the ABA signal (KF Mayer et al. 2012; Gosti et al. 1999; Merlot et al. 2001). In this study, the eQTL for amy1 at DPA30 was located close to the centromere of chromosome 3DS, approximately 35 cM from TaVP-1D. A minor QTL associated with PHS has been reported on chromosome 3DS (Kulwal et al. 2004). Use of linkage markers from a previously constructed consensus map (Somers et al. 2004) indicated that the eQTL on chromosome 3DS (eqamy1-DPA30.3D) is probably in the vicinity of the QTL for PHS. This suggests that this region plays a role in the regulation of PHS through effecting amy1 expression. Among the above genes, Vp1 is known to repress downstream amylase in developing seeds of maize and wheat (Hoecker et al. 1999; Utsugi et al. 2008), while the other genes could directly or indirectly interact with Vp1/ABI3 in ABA signaling (Lopez-Molina et al. 2002; Gosti et al. 1999; Kurup et al. 2000). This evidence suggests that the eQTL on chromosome 3DS may be downstream of the ABA signal; further study is required to prove this prediction.
Two eQTLs for amy1, eqamy1-DPA25.1B and eqamy1-DPA30.1B, located close to the centromere of chromosome 1B were detected at DPA25 and DPA30, respectively. They were in the same domain, positioned 7 cM from the centromere on the abbreviated map, and both had a negative effect on amy1 mRNA accumulation. Previously, a minor QTL associated with PHS was detected on chromosome 1B, this is close to the eQTLs region reported in this study (Munkvold et al. 2009). Because genes involved in PHS have rarely been found on group 1 chromosomes, further detection analysis is required to characterize these two regions upstream of amy1.
Recently, quantity trait analysis has aimed at detecting large effect QTLs. These QTLs exist upstream of signal transduction processes, with pleiotropic effects controlling target traits (Rasul et al. 2009; Singh et al. 2007; Zhang et al. 2011; Li et al. 2012; Lin et al. 2006). In the present study, amy1 encoded a downstream enzyme regulated by unknown transcriptional factors. Regions affecting amy1 expression have been represented as minor QTLs for PHS in other studies, suggesting that minor QTLs affect target traits though either single or minor ways. Molecular characterization of both loci would give insight into the complex mechanisms of PHS. Recently, many QTLs and linkage markers have elucidated PHS. Although genetic improvements may accelerate wheat selection, a detailed understanding of the complex network of QTLs or genes for seed traits and plant development physiology will promote successful molecular breeding programs. The use of traditional QTL methodologies alone will not be enough to reach this goal, and eQTL analysis of candidate genes will be of great importance for explaining the complex architecture of PHS and may even detect new loci. Therefore, as with QTLs, significant eQTLs are likely to be incorporated into maker-assisted selection wheat breeding programs.
References
Appleford NE, Wilkinson MD, Ma Q, Evans DJ, Stone MC, Pearce SP, Lenton JR (2007) Decreased shoot stature and grain α-amylase activity following ectopic expression of a gibberellin 2-oxidase gene in transgenic wheat. J Exp Bot 58(12):3213–3226
Bailey PC, McKibbin RS, Lenton JR, Holdsworth MJ, Flintham JE, Gale MD (1999) Genetic map locations for orthologous Vp1 genes in wheat and rice. Theor Appl Genet 98(2):281–284
Barrero JM, Mrva K, Talbot MJ, White RG, Taylor J, Gubler F, Mares DJ (2013) Genetic, hormonal, and physiological analysis of late maturity α-amylase in wheat. Plant Physiol 161(3):1265–1277
Baulcombe DC, Huttly AK, Martienssen RA, Barker RF, Jarvis MG (1987) A novel wheat α-amylase gene (α-Amy3). Mol Gen Genet 209(1):33–40
Buchanan AM, Nicholas EM (1980) Sprouting, alpha-amylase, and breadmaking quality. Cereal Res Commun 8(1):23–28
Chen X, Hackett CA, Niks RE, Hedley PE, Booth C, Druka A, Waugh R (2010) An eQTL analysis of partial resistance to Puccinia hordei in barley. PLoS ONE 5(1):e8598
Cheng C-R, Oldach K, Mrva K, Mares D (2013) Analysis of high pI a-Amy-1 gene family members expressed in late maturity a-amylase in wheat (Triticum aestivum L.). Mol Breeding 33(3):519–529
Daussant J, Renard HA (1987) Development of different α-amylase isozymes, having high and low isoelectric points, during early stages of kernel development in wheat. J Cereal Sci 5(1):13–21
De Laethauwer S, De Riek J, Stals I, Reheul D, Haesaert G (2013) α-Amylase gene expression during kernel development in relation to pre-harvest sprouting in wheat and triticale. Acta Physiol Plant 35:2927–2938
Derera NF (1989) The effects of preharvest rain. In: Derera NF (ed) Preharvest sprouting in cereals. CRC Press Inc, Boca Raton, pp 2–14
Fedorova L, Fedorov A (2003) Introns in gene evolution. In: Origin and evolution of new gene functions. Springer, The Netherlands, pp 123–131
Fofana B, Humphreys DG, Rasul G, Cloutier S, Brûlé-Babel A, Woods S, Somers DJ (2009) Mapping quantitative trait loci controlling pre-harvest sprouting resistance in a red × white seeded spring wheat cross. Euphytica 165(3):509–521
Gale MD, Ainsworth CC (1984) The relationship between α-amylase species found in developing and germinating wheat grain. Biochem Genet 22(11):1031–1036
Gale MD, Law CN, Chojecki AJ, Kempton RA (1983) Genetic control of α-amylase production in wheat. Theor Appl Genet 64(4):309–316
Garg K, Green P, Nickerson DA (1999) Identification of candidate coding region single nucleotide polymorphisms in 165 human genes using assembled expressed sequence tags. Genome Res 9:1087–1092
Gómez-Cadenas A, Zentella R, Sutliff TD, Ho THD (2001) Involvement of multiple cis-elements in the regulation of GA responsive promoters: definition of a new cis-element in the Amy32b gene promoter of barley (Hordeum vulgare). Physiol Plant 112(2):211–216
Gosti F, Beaudoin N, Serizet C, Webb AA, Vartanian N, Giraudat J (1999) ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. Plant Cell 11(10):1897–1910
Higgins TJ, Jacobsen JV, Zwar JA (1982) Gibberellic acid and abscisic acid modulate protein synthesis and mRNA levels in barley aleurone layers. Plant Mol Biol 1(3):191–215
Hoecker U, Vasil IK, McCarty DR (1999) Signaling from the embryo conditions Vp1-mediated repression of α-amylase genes in the aleurone of developing maize seeds. Plant J 19(4):371–377
Huang N, Stebbins GL, Rodriguez RL (1992) Classification and evolution of alpha-amylase genes in plants. Proc Natl Acad Sci 89(16):7526–7530
Huttly AK, Martienssen RA, Baulcombe DC (1988) Sequence heterogeneity and differential expression of the α-Amy2 gene family in wheat. Mol Gen Genet 214(2):232–240
Huttly AK, Phillips AL, Tregear JW (1992) Localisation of cis elements in the promoter of a wheat α-Amy2 gene. Plant Mol Biol 19(6):903–911
Jansen RC, Nap JP (2001) Genetical genomics: the added value from segregation. Trends Genet 17(7):388–391
Jordan MC, Somers DJ, Banks TW (2007) Identifying regions of the wheat genome controlling seed development by mapping expression quantitative trait loci. Plant Biotechnol J 5(3):442–453
Kadziola A, Abe JI, Svensson B, Haser R (1994) Crystal and molecular structure of barley α-amylase. J Mol Biol 239(1):104–121
Knox CAP, Sonthayanon B, Chandra GR, Muthukrishnan S (1987) Structure and organization of two divergent α-amylase genes from barley. Plant Mol Biol 9:3–17
Kruger JE (1976) Biochemistry of pre harvest sprouting in cereals and practical applications in plant breeding. Cereal Res Commun 4:187–194
Kulwal PL, Singh R, Balyan HS, Gupta PK (2004) Genetic basis of pre-harvest sprouting tolerance using single-locus and two-locus QTL analyses in bread wheat. Funct Integr Genomics 4(2):94–101
Kurup S, Jones HD, Holdsworth MJ (2000) Interactions of the developmental regulator ABI3 with proteins identified from developing Arabidopsis seeds. Plant J 21(2):143–155
Lazarus CM, Baulcombe DC, Martienssen RA (1985) α-Amylase genes of wheat are two multigene families which are differentially expressed. Plant Mol Biol 5(1):13–24
Li S, Wang C, Chang X, Jing R (2012) Genetic dissection of developmental behavior of grain weight in wheat under diverse temperature and water regimes. Genetica 140(7–9):393–405
Lin F, Xue SL, Zhang ZZ, Zhang CQ, Kong ZX, Yao GQ, Ma ZQ (2006) Mapping QTL associated with resistance to Fusarium head blight in the Nanda2419 × Wangshuibai population. II: type I resistance. Theor Appl Genet 112(3):528–535
Long XY, Wang JR, Ouellet T, Rocheleau H, Wei YM, Pu ZE, Zheng YL (2010) Genome-wide identification and evaluation of novel internal control genes for Q-PCR based transcript normalization in wheat. Plant Mol Biol 74(3):307–311
Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH (2002) ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J 32(3):317–328
MacGregor AW, Dushnicky L (1989) α-Amylase in developing barley kernels-a reappraisal. J Inst Brew 95(1):29–33
Mares D, Mrva K (2008) Late-maturity-amylase: low falling number in wheat in the absence of preharvest sprouting. J Cereal Sci 47(1):6–17
Mares DJ, Oettler G (1991) α-Amylase activity in developing triticale grains. J Cereal Sci 13(2):151–160
Masojć P, Milczarski P (2009) Relationship between QTLs for preharvest sprouting and alpha-amylase activity in rye grain. Mol Breed 23(1):75–84
Mayer KF, Waugh R, Brown JW, Schulman A, Langridge P, Platzer M, Stein N (2012) A physical, genetic and functional sequence assembly of the barley genome. Nature 491(7426):711–716
Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J (2001) The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. Plant J 25(3):295–303
Munkvold JD, Tanaka J, Benscher D, Sorrells ME (2009) Mapping quantitative trait loci for preharvest sprouting resistance in white wheat. Theor Appl Genet 119(7):1223–1235
Munkvold JD, Laudencia-Chingcuanco D, Sorrells ME (2013) Systems genetics of environmental response in the mature wheat embryo. Genetics 194(1):265–277
Nakamura S, Toyama T (2001) Isolation of a VP1 homologue from wheat and analysis of its expression in embryos of dormant and non-dormant cultivars. J Exp Bot 52(357):875–876
Nakamura S, Komatsuda T, Miura H (2007) Mapping diploid wheat homologues of Arabidopsis seed ABA signaling genes and QTLs for seed dormancy. Theor Appl Genet 114(7):1129–1139
Nakamura S, Abe F, Kawahigashi H, Nakazono K, Tagiri A, Matsumoto T, Miura H (2011) A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. Plant Cell Online 23(9):3215–3229
Osa M, Kato K, Mori M, Shindo C, Torada A, Miura H (2003) Mapping QTLs for seed dormancy and the Vp1 homologue on chromosome 3A in wheat. Theor Appl Genet 106(8):1491–1496
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res 29(9):e45
Potokina E, Druka A, Luo Z, Wise R, Waugh R, Kearsey M (2008) Gene expression quantitative trait locus analysis of 16 000 barley genes reveals a complex pattern of genome-wide transcriptional regulation. Plant J 53(1):90–101
Rasul G, Humphreys DG, Brule-Babel A, McCartney CA, Knox RE, DePauw RM, Somers DJ (2009) Mapping QTLs for pre-harvest sprouting traits in the spring wheat cross ‘RL4452/AC Domain’. Euphytica 168(3):363–378
Rodenburg KW, Juge N, Guo XJ, Søgaard M, Chaix JC, Svensson B (1994) Domain B protruding at the third β strand of the α/β barrel in barley α-amylase confers distinct isozyme-specific properties. Eur J Biochem 221(1):277–284
Sargeant JG (1980) alpha-Amylase isoenzymes and starch degradation. Cereal Res Commun 8(1):77–86
Singh S, Sharma I, Sehgal SK, Bains NS, Guo Z, Nelson JC, Bowden RL (2007) Molecular mapping of QTLs for Karnal bunt resistance in two recombinant inbred populations of bread wheat. Theor Appl Genet 116(1):147–154
Somers DJ, Isaac P, Edwards K (2004) A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109(6):1105–1114
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24(8):1596–1599
Tregear JW, Primavesi LF, Huttly AK (1995) Functional analysis of linker insertions and point mutations in the α-Amy2/54 GA-regulated promoter. Plant Mol Biol 29(4):749–758
Utsugi S, Nakamura S, Noda K, Maekawa M (2008) Structural and functional properties of Viviparous1 genes in dormant wheat. Genes Genet Syst 83(2):153–166
Varughese G, Barker T, Saari E (1986) Triticale CIMMYT, Mexico, DF. ISBN 968-6127-11-9
Vergne E, Grand X, Ballini E, Chalvon V, Saindrenan P, Tharreau D, Morel JB (2010) Preformed expression of defense is a hallmark of partial resistance to rice blast fungal pathogen Magnaporthe oryzae. BMC Plant Biol 10(1):206
Wang S, Basten CJ, Zeng ZB (2007) Windows QTL cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC
Wang J, Yu H, Xie W, Xing Y, Yu S, Xu C, Zhang Q (2010) A global analysis of QTLs for expression variations in rice shoots at the early seedling stage. Plant J 63(6):1063–1074
Yang Y, Ma YZ, Xu ZS, Chen XM, He ZH, Yu Z, Xia LQ (2007) Isolation and characterization of Viviparous-1 genes in wheat cultivars with distinct ABA sensitivity and pre-harvest sprouting tolerance. J Exp Bot 58(11):2863–2871
Yu M, Chen GY (2013) Conditional QTL mapping for waterlogging tolerance in two RILs populations of wheat. Springer Plus 2(1):1–7
Zhang J, Hao C, Ren Q, Chang X, Liu G, Jing R (2011) Association mapping of dynamic developmental plant height in common wheat. Planta 234(5):891–902
Acknowledgments
This work was supported by the National Basic Research Program of China (2014CB147200) and the National Natural Science Foundation of China (31171555, 31230053). JR Wang was supported by Fok Ying Tong Education Foundation (131023).
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Yang, J., Liu, Y., Pu, Z. et al. Molecular characterization of high pI α-amylase and its expression QTL analysis in synthetic wheat RILs. Mol Breeding 34, 1075–1085 (2014). https://doi.org/10.1007/s11032-014-0098-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-014-0098-z