Abstract
The root proteome of nitrogen-efficient and nitrogen-inefficient rice cultivars was compared in this study in order to investigate the differential expression of proteins under deficient (1 mM), low (10 mM) and high (25 mM) levels of nitrogen (N). Nitrogen use efficiency (NUE) was assessed by biochemical assays such as N-uptake kinetics and activities of N-assimilation enzymes. Two-dimensional gel electrophoresis and MALDI–TOF–MS analysis resulted in the identification of 504 protein spots (210 and 294 spots in cvs. Rai Sudha and Munga Phool, respectively). A positive correlation was observed between physiological parameters and the concentration of a number of root proteins. Sixty-three spots showed a significant cultivar × N-treatment effect on the level of expression. Functional aspects of eleven spots with major alterations in expression over control were critically analyzed. The data suggest that glutamine synthetase, cysteine proteinase inhibitor-I, porphobilinogen deaminase (fragment) and ferritin were involved in conferring N efficiency to the N-efficient rice cultivars/genotypes. Interestingly, these proteins are involved directly or indirectly in N assimilation. Such studies should help us in identifying and understanding the structural or functional protein(s) involved in the response to the level of nitrogen fertilization.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Nitrogen (N), an essential macronutrient for plant growth and yield, helps in increasing the production of food to feed the exponentially increasing human population. On a global basis, about 40 % of the human population relies on N fertilizer for food production (Smil 1999). Of the cereal crops, rice is consumed by more than half of the world’s population. However, it utilizes only 30–40 % of total N applied (Abrol et al. 1999). The unused 60–70 % nitrogen poses severe environmental as well as human health threats (Anjana and Iqbal 2007). In such a condition, it becomes important to search for cultivars/genotypes that may have only a limited yield decline in response to N deficiency (Hirel et al. 2005), i.e., the N-efficient cultivars/genotypes capable of yielding well even under N-deficient conditions. The N-efficiency mechanism depends upon various signal molecules present in the shoot and roots. Most of the proteomic research done so far has focused on shoots of cereal crops, though their roots also deserve attention. Roots possess specific transporters for the intake of nutrients, while leaves are the site for nitrogen assimilation. Plants take up nitrogen in the form of nitrate from the soil through transporter systems located in the roots. After uptake, nitrate (NO3 −) is assimilated by a series of nitrate assimilatory enzymes, viz., nitrate reductase (NR), nitrite reductase (NiR), glutamine synthetase (GS) and glutamate synthase (GOGAT). Nitrate reductase catalyzes the first step of nitrate assimilation in plants, leading to reduction of nitrate to nitrite with pyrimidine nucleotide in higher plants (Lea and Ireland 1999). Acting as a catalyst for conversion of amino acid glutamate into amide, glutamine synthetase also has a role in controlling plant nitrogen assimilation (Hakeem et al. 2011). The effect of nitrogen starvation on root is evident from the N-uptake rate and the total biomass accumulation. Considering two cultivars of the same crop with differential nitrogen use efficiency (NUE) should to a large extent rule out variation in the anatomy and physiological chemistry of roots.
In the recent era of genomics, genetic manipulation of key genes responsible for N uptake and assimilation is one of the basic approaches for enhancing NUE. However, our knowledge concerning gene expression and regulation of plant response to low N, so frequently met with in agricultural field conditions, is still deficient (Pathak et al. 2008). It is essential to develop strategies for manipulating the genetic architecture of plants to improve their NUE, and also to identify the signal transduction pathways and the elements that function to regulate genes involved in N uptake and assimilation. Since signal transduction and gene regulation are based on proteins at large, it is germane to focus on protein-expression pattern. Furthermore, a research tool is needed to identify and understand the role of various proteins at a given point in time. Proteomics is a high-throughput biotechnological approach being used to understand the biological function of proteins in response to different biotic or abiotic stresses (Agarwal et al. 2002; Kim et al. 2004). Quantitative as well as qualitative differential expression of protein spots may help in identifying the essential molecules (enzymes) responsible for N uptake and assimilation, which may explain the cause of high N efficiency in cereal crops.
The present study screened twenty cultivars (cvs.) of rice, commonly grown in India, for their NUE, as determined by biochemical assays. Two rice cultivars, showing the maximum (cv. Rai Sudha) and minimum (cv. Munga Phool) NUE, were then grown hydroponically with three [low (1 mM), moderate (10 mM) and high (25 mM)] levels of N supply for 20 days in order to harvest their roots for proteomic studies. Two-dimensional gel electrophoresis and MALDI–TOF–MS were employed for proteomic maps and protein identification, respectively. Studies aimed at identifying and understanding the proteins responding to different levels of N fertilization must help in improving NUE among crop plants.
Materials and methods
Plant material and growth conditions
Twenty accessions of rice (Oryza sativa L.) cultivars commonly grown at geographically different sites (northern and eastern parts of India) were chosen for this study. These genotypes, procured from Sher-e-Kashmir University of Agricultural Sciences and Technology (Srinagar, India), GB Pant University (Pant Nagar, India) and Gene Campaign (Ranchi, India), were designated G1–G20 (Table 1). Seeds were washed with detergent and surface-sterilized, using 1 % sodium hypochlorite, for 10 min, followed by 10 washings with double-distilled water (DDW). These were then germinated in a fine net tied on the top of 250 ml beakers containing DDW and covered with few layers of moist cheese-cloth. Germinated seedlings of similar size (5 days old) were supplied with 250 ml of nutrient solution containing 5.6 mM K+, 3.4 mM Ca2+, 0.9 mM Mg2+, 0.9 mM PO4 −, 21.5 μM Fe2+, 23 μM B3+, 9 μM Mn2+, 0.30 μM Mo2+, 0.90 μM Cu2+ and 3.50 μM Zn2+, replacing the DDW in the hydroponics culture system. The nutrient solution was aerated using aquarium air pumps, the nutrient solution being replaced every third day and continued for 20 days. The experiment was conducted with five biological replicates (10 plants per replicate). Nitrogen was supplied in the form of nitrate with three concentrations of KNO3 [deficient (1 mM), low (10 mM) and high (25 mM), hereafter referred as T0, T1 and T2, respectively]. The growth chamber was maintained at a photosynthetic photon flux density of 430 μmol m−2 s−1, 14/10-h light/dark period and a relative humidity of 60 %.
Physiochemical parameters
Nitrogen efficiency
Seedlings (30 days old) from each treatment were washed, weighed and dried in a hot air oven at 65 ± 2 °C for 72 h to determine biomass or dry weight (DW) accumulation. Nitrogen efficiency (NE) was calculated as follows (Ahmad et al. 2008):
Nitrate uptake kinetics
Nitrate uptake was measured in 10-day-old seedlings in two sets of different levels of nitrate supply, viz. (a) a high external nitrate range (10, 15, 25, 50, 75 and 100 μM KNO3) and (b) a low external nitrate range (10, 15, 25, 50, 75 and 100 nM KNO3) for 5 h. The nitrate uptake rates were determined from the linear regression of nitrate depleted from the solution due to uptake, which represents the net flux into the plant through roots. All ion fluxes were correlated to the fresh weight of the root tissue used during the uptake studies. Nitrate ions in these samples were measured by high-performance liquid chromatography (HPLC, Waters, USA), using 10 mm × 250 mm analytical anion exchange column (Whatman Partisil-10 SAX), according to Hunt and Seymour (1985). The UV detector was used at 210 nm. Sodium phosphate buffer (50 mM/pH 8) was used as eluent and the flow rate was kept at 1 ml/min with 25 μl of sample injection volume. The run time was maintained at 15 min. The K m and V max values were computed by using the Lineweaver–Burk plot of 1/V against 1/[S], where V and [S] represent velocity of uptake and substrate concentration, respectively. The values were expressed in mM NO3 − and μM NO3 − [g fresh weight (FW) of root]−1 min−1, respectively.
In-vitro nitrate reductase activity
In-vitro nitrate reductase activity (NRA) was analyzed by the method of Ahmad and Abdin (1999). Root samples (1 g FW) were frozen in liquid nitrogen and homogenized in 250 ml extraction medium consisting of 25 mM potassium phosphate buffer (pH 7.5). The extract was filtered through muslin cloth and then centrifuged. This crude extract was assayed for enzymatic activity. The enzyme extract and reaction mixture containing 0.5 ml of 100 μM potassium phosphate buffer pH 6.8 was incubated at 33 °C for 30 min. The reaction was stopped by the addition of zinc acetate. Nitrite produced was estimated by the method of Evans and Nason (1953) at 540 nm using ultraviolet (UV)–Vis spectrophotometer (Model λ-bio-20, PerkinElmer, Germany). The enzyme activity was expressed as μmol g−1 DW h−1. The experiment was done five times for each sample with three independent extractions, carried out for each of the different assays.
Glutamine synthatase activity
Root tissue (1 g FW) was extracted in extraction medium (in the ratio 1:3) containing 25 mM Tris–HCl buffer (pH 7.6), 1 mM MgCl2, 10 mM β-mercaptoethanol and 1 mM DTT (McNally et al. 1983). The extracts were centrifuged at 20,000g for 30 min at 4 °C and the supernatant was used immediately for the glutamine synthase (GS) assay, following the method of Rhodes et al. (1975). The reaction mixture (in a final volume of 3 ml) contained 100 μmol Tris–HCl buffer (pH 8), 10 μmol ATP, 250 μmol sodium glutamate, 100 μmol MgSO4, 10 μmol l-cysteine, 10 μmol hydroxylamine and enzyme extract. In order to start the reaction, hydroxylamine was added to the mixture, which was then incubated for 30 min at 30 °C. Formation of γ-glutamyl hydroxamate (γ-GHA) was determined by adding 1 ml of ferric chloride reagent. Absorbance was recorded at 540 nm. Enzyme activity was expressed in μmol γ-GHA g−1 DW min−1. The experiment was done five times for each sample with three independent extractions, carried out for each of the different assays.
Soluble protein content
For quantification of soluble protein, 0.5 g of fresh leaf material was homogenized in 1.5 ml of extraction buffer at 4 °C. Protein content was determined according to Bradford (1976) using bovine serum albumin as a standard. The homogenate was transferred to a 2-ml tube and centrifuged at 5,000g for 10 min at 4 °C. An equal amount of chilled trichloroacetic acid (TCA) (10 %) was added to the supernatant, which was again centrifuged at 3300 g for 10 min. The supernatant was discarded and the pellet was washed with acetone and then dissolved in 1 ml of 0.1 N NaOH. The absorbance was recorded at 595 nm on a UV–Vis spectrophotometer (Bio 20, PerkinElmer, Germany). The soluble protein content was calculated with the help of a standard curve of bovine albumin serum (Sigma, USA) and the content was expressed in mg g−1 DW. The experiment was done five times for each sample with three independent extractions, carried out for each of the different assays.
Proteomic study
Protein extraction from roots
Frozen samples of root were ground in a chilled mortar and pestle in liquid nitrogen and incubated overnight with chilled (−20 °C) acetone solution containing 12.5 % (w/v) TCA and 0.07 % (v/v) β-mercaptoethanol (Natarajan et al. 2005). The precipitated proteins were centrifuged to form a pellet at 12,000g along with cellular debris and washed five times, incubating with chilled acetone mixture for 1 h between each step. The last step of washing involved only acetone and overnight incubation at −20 °C. The resultant was centrifuged at 6,000g for 10 min and the white pellet thus obtained was vacuum-dried, re-suspended in resolubilization solution (9 M urea, 1 % CHAPS, 1 % DTT, 1 % pharmalyte), sonicated at 4 °C for 5 min and mixed vigorously at room temperature for 1 h. The mixture was centrifuged 21,000g at 4 °C at for 30 min. The pellet was discarded and the supernatant retained was analyzed for protein concentration, using Bradford reagent (Bradford 1976) and the bovine serum albumin standard curve.
2D-PAGE
Protean IEF Cells (Bio-Rad, USA) were used for isoelectric focusing (IEF) of protein in immobilized pH gradient (IPG) strips (Bio-Rad; pH 4.0–7.0, linear gradient, 11 cm). The IPG strips were rehydrated for 12 h with 250 μl rehydration buffer (8 M urea, 2 M thiourea, 2 % CHAPS, 0.5 % Pharmalyte, 0.002 % bromophenol blue) containing 350 μg proteins. The voltage programme for the IEF run was 500 V for 1 h, 1,000 V for 1 h, 2,000 V for 1 h, and 3,000 V for the rest of the time to reach a total of 35 kVh. Initially, the current was kept at 50 μA, which changed automatically with decreasing resistance. Following the IEF run, the IPG strips, containing focused protein, were treated with 1 % dithioerythritol, followed by 2.5 % indoleacetic acid for 15 min, each prepared separately in equilibration buffer consisting of 50 mM Tris–HCl pH 8.8, 6 M urea, 30 % glycerol, 2 % sodium dodecyl sulfate (SDS) and 0.002 % bromophenol blue. The two-dimensional (2D) run was performed with IPG strips on a 12.5 % acrylamide gel, using a Mini Tetrad electrophoretic unit (Bio-Rad) at 100 V. The 2D gels were stained with Coomassie Brilliant Blue (CBB G-250) stain as mentioned by Newsholme et al. (2000). Following SDS electrophoresis, the gels were fixed overnight in 1.5 l of 50 % ethanol/3 % phosphoric acid per 10 gels and then washed three times for 30 min in 1.5 l of cold double ionized water per 10 gels. They were transferred to 1.5 l of 34 % methanol/17 % aluminium sulfate/3 % phosphoric acid per 10 gels for 1 h. After the addition of 1 g powdered CBB G-250 the gels were stained for 3 days to achieve equilibrium in intensity. The gels so developed were scanned and digitized using a calibrated imaging densitometer (GS 710, Bio-Rad). The pattern of protein distribution on 2D gels was compared with the help of PD-Quest software (Bio-Rad). The intensity was quantified for each spot in each gel, using Dunnett’s multiple range test and taking p < 0.05 as significant. At least three replicates (both biological as well as technical) were performed for each sample.
In-gel digestion of protein spots
Protein spots of interest were excised carefully from the 2D gels, with each spot in a separate micro-tube, and protein digestion was performed as described by Qureshi et al. (2010). The excised gel pieces containing protein were washed with CH3CN:H2O (1:1, v/v), containing 25 mM ammonium bicarbonate, to remove the blue stain. The gel pieces were dehydrated with 100 % acetonitrile, dried under vacuum and incubated overnight at 37 °C with 20 μl of 10 μg/ml porcine trypsin prepared in 20 mM ammonium bicarbonate. The resulting tryptic fragments were eluted by diffusion into CH3CN:H2O and 0.5 % trifluoroacetic acid (1:1, v/v). A sonic bath was used to facilitate the diffusion. The extract was vacuum-dried and the pellet was dissolved in CH3CN:H2O and 0.1 % trifluoroacetic acid (1:1, v/v).
Mass spectrometry
For peptide mass fingerprinting (PMF), a Voyager DE-STR MALDI–TOF mass spectrometer (Model 4800, Applied Biosystems, UK) operating in positive ion reflector mode was used to analyze the tryptic peptides. Samples were co-crystallized with α-cyanohydroxycinnamic acid (CHCA) matrix, and spectra were acquired with 400 shots of a 337 nm nitrogen laser operating at 20 Hz. The spectra were calibrated using the trypsin autolysis peaks at m/z 842.51 and 2.211.10 as internal standards. The raw data were processed by Sequest to generate DTA files for database searching. The ‘merge pl’ script from Matrix Science was used to convert multiple Sequest DTA files into a single Mascot generic file suitable for searching in Mascot.
PMF data analysis
Protein identification was performed using the Mascot search engine (http://www.matrixscience.com), which uses a probability-based scoring system (Perkins et al. 1999). NCBI non-redundant and SwissProt databases were selected as the primary databases to be searched for protein sequence matches from the Viridiplantae taxon. The parameters for database search with MALDI–TOF PMF data and with MS/MS spectra were set as previously (Song et al. 2007). For MALDI–TOF–MS data to qualify as a positive identification, a protein’s score had to equal or exceed the minimum significant score (95 % significance threshold; p ≥ 0.05) of 64 for NCBInr or 55 for SwissProt database searching.
Statistical analysis
Values are expressed as mean ± SEM. Differences between treatments were analyzed by one-way ANOVA, taking p < 0.05 as significant, according to Dunnett’s multiple range test.
Results and discussion
Growth and physiological parameters
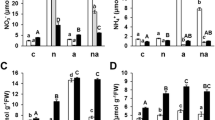
In the present study, rice plants were grown hydroponically. Whilst it is recognized that this will have some effect on protein expression, so altering the proteome, this limitation is considered to be acceptable (Conn et al. 2013; Hakeem et al. 2012a, b; Khan et al. 2013; Peng et al. 2009; Wang et al. 2012). Nitrogen availability can be more tightly controlled when delivered in a hydroponic medium and, importantly, it prevents the roots from being damaged when harvested; such damage would also effect the proteome, particularly stress response genes. The 20 rice cultivars screened for their nitrogen efficiency (NE) exhibited a huge difference in NE (30–95.7 % at T1 = 10 mM and 38.53–93.5 % at T2 = 25 mM respectively) (Table 1). The NE calculated on the basis of dry weight of the whole shoot is frequently used to assess genotypic variation in tolerance to nutrient deficiency (Ahmad et al. 2005; Fageria and Baligar 2005; Chandna et al. 2010). Considering the whole shoot growth, tolerance to N deficiency (relative growth) was determined with the help of the calculated NE. On the basis of differential NE of rice cultivars, these cultivars were grouped into Low Nitrogen Efficient (LNE), Moderate Nitrogen Efficient (MNE) and High Nitrogen Efficient (HNE) categories. Cv. Rai Sudha was thus identified as a highly N-use-efficient cultivar (HNE), while cv. Munga Phool was identified as the most N-use-inefficient cultivar (LNE) of rice (Table 1).
In Rai Sudha, increase in the level of N treatment (10 and 25 mM) did not affect its NR and GS activities (Tables 2, 3) or its soluble protein content (Supplemental Table 4). On the other hand, in Munga Phool, the NR and GS activities as well as the soluble protein content increased significantly with increase in N treatment. The NR activity of Munga Phool showed a significant increase of 54 % with T1 (10 mM N treatment), and of 80 % with T2 (25 mM N treatment). NR could play an indirect role in the absorption of nitrate, regulating the levels of nitrate and amino acids in root cells. Since nitrate induces nitrate reductase, the rate of nitrate uptake to the site of induction is the main controlling factor for NR activity. In our experiment, the pattern of nitrate uptake by roots as well as the nitrate and nitrogen contents in roots coincided with the pattern of NR activity. The difference in NR activity was possibly connected to differences in the contents of nitrate ions in the tissue. GS activity in Munga Phool increased by 92 % with T1 and 169 % with T2, in comparison to T0 (1 mM N). It was the maximum in HNE cultivars at T0. However, no significant effect was observed with increase in N supply, showing that HNE genotypes were able to utilize the absorbed N and thus grow well at low N. The MNE cultivars showed increase in GS activity with increase in N supply at T1. With further increase in N supply i.e., at T2 (25 mM), no significant change was observed. In the LNE cultivars, a significant increase in GS activity was noted with increase in N supply (Table 3). Previous studies have revealed that in the LNE genotypes a substantial portion of N is not re-translocated to harvested structures and the GS activity in leaves is lower than in the HNE genotypes (Habash et al. 2001; Masclaux et al. 2001). Hence, the ability of the HNE genotypes to harvest more N and redistribute it to grains is due to their well-coordinated system of N uptake and assimilation. In LNE cultivars, GS activity was sufficiently high to maintain a low level of ammonium in leaf tissues irrespective of the external N supply (Cao et al. 2008); this explains the increase in activity of GS with increase in N supply. There was a significant effect of 10 and 25 mM levels of N on the nitrogen content of Munga Phool. An increase of 85 and 152 % was caused by the supply of 10 mM and 25 mM N respectively, as compared with T0 (1 mM N). The increase was 68 % with T2 over T1. The N content of both the cultivars remained unaffected by the high N levels, compared with the control (Supplemental Table 5).
Since it is the final product that demonstrates the efficiency of any system, we estimated the soluble protein content of different cultivars to determine the efficiency of their N cycle. In the LNE cultivars (Munga Phool), a significant increase in protein content was observed with both T1 (10 mM N) and T2 (25 mM N), in comparison with T0 (Supplemental Table 4). However, these treatments did not affect the soluble protein content of HNE cultivars. Bahrman et al. (2005) determined the differential changes in root-protein patterns in two winter varieties of wheat under high and low N nutrition levels. Smith (1980) found a decline in the protein content of cultured tobacco cells due to N starvation. In our study, the soluble protein content was significantly less in roots than in leaves in all the cultivars with all the N treatments (Hakeem et al. 2011).
Root uptake kinetics
The amount of nitrate depleted from the nutrient solutions was taken to represent the nitrate uptake. The K m value, ranging from 0.0075 to 0.052 mM, was the lowest in cv. Rai Sudha and highest in cv. Munga Phool. In contrast, the V max values (0.8–4.8 μmol [g root FW]−1 min−1) were minimum in Munga Phool and maximum in Rai Sudha (Supplemental Table 6). Studies on the kinetic and energetic aspects of nitrate uptake are confined mostly to low-concentration systems. In the low concentration range, the net nitrate uptake rate normally obeys Michaelis–Menten kinetics (Clarkson 1985). As for the kinetic parameters V max and K m, it is rather difficult to relate the magnitude of cultivar difference to the performance under nutrient-limiting conditions. In the present study, the K m values of the twenty cultivars varied from 0.0075 to 0.052 mM, showing similarity with other species ranging from 0.007 to 0.187 mM (Goyal and Huffaker 1986; Aslam et al. 1992; Carezo et al. 1997). Woodend et al. (1986) observed a 4-fold difference of apparent K m among eight wheat varieties studied. A K m value of 7.5 mM has been reported for excised corn root (Neyra and Hageman 1976). Variations in K m values can possibly be explained on the basis of endogenous nitrate levels of plants, molybdenum (Mo) status of roots, and intensive breeding of certain species for high nitrogen response. Moreover, since nitrate efflux by roots is affected by their nitrate content, factors that affect nitrate reduction during plant growth should also affect the net nitrate uptake rate and its K m.
V max (i.e., the capacity of the saturable component of the uptake system) also varied significantly among the twenty rice genotypes. The values ranged from 0.8 to 4.8 μmol [g root FW]−1 min−1. Barley cultivars have shown significant variation in net nitrate uptake rate, which was greater among accessions than among species (Bloom 1985). Inter-genotype variation in the quantity (number) of transport molecules per unit of the absorbing cell membrane possibly provides a theoretical explanation for differences in V max (Pace and McClure 1986).
Proteomics study
The rice root protein pattern obtained by 2D gel electrophoresis depicted 210 spots in cv. Rai Sudha and 294 spots in cv. Munga Phool grown under different N treatments. The intensity of 63 protein spots changed markedly in both the genotypes by N treatments (T1 and T2), by comparison to T0. Nineteen spots (nine in Rai Sudha and ten in Munga Phool) showed variations, whereas 13 spots (four in Rai Sudha and nine in Munga Phool) remained unaffected by the N treatment. The variation of protein-spot intensity due to N nutrition level of the two cultivars is presented in Supplemental Table 7. Our observations conform to those of Bahrman et al. (2005) on wheat varieties. We analyzed a total of 84 protein spots quantitatively as well as qualitatively. Forty-three spots were identified through Mascot analysis and eleven of them were critically analyzed for functional aspects (Fig. 1). Most of the spots were related to metabolism and stress responses (60 %) (Supplemental Table 8), suggesting that N levels directly influence the different metabolic pathways of a plant. A significant cultivar × treatment interaction was found for nitrate concentration with both Rai Sudha and Munga Phool at low N level. Among the identified root spots in Rai Sudha, the intensity of nine spots (1, 2, 3, 4, 5, 7, 8, 10 and 11) increased with increase in the N applied. Spot 9 showed increased intensity at T1 by comparison with T0, but the intensity again decreased at T2. However, one spot (6) did not show any change with increasing levels of N (Fig. 2). Figure 2 shows the expression level of 11 protein spots from root-map gels of Rai Sudha and Munga Phool, which were successfully identified by MALDI–TOF or MS/MS analysis and Mascot search, and whose expression was altered differentially at different N levels. At T0 (1 mM N), the expression level of all the 11 spots was highly variability among cultivars. Spots 3, 6, 10 and 11 were more highly expressed in Rai Sudha than in Munga Phool, whereas the rest were more significant in Munga Phool than in Rai Sudha. At T1 (10 mM N), the expression level of all the protein spots in Rai Sudha was significantly increased. On the whole, a 2–5-fold increase in expression was observed in Rai Sudha at T1, in comparison with T0.
Differential changes in root proteomic profile of rice cultivars Rai Sudha (N-efficient genotype) and Munga Phool (N-inefficient) exposed to low (T1, 10 mM N) and high (T2, 25 mM N) N treatments for 20 days. Proteins were extracted from roots, separated by 2D-PAGE and stained by CBB. Following scanning, the gel patterns were analyzed using PD-Quest software. Numbered spots (differentially expressed) were selected for MS. Areas of gels with significant differential expression are shown. Intensity was quantified for each spot in each gel, using Dunnett’s multiple range test and taking p < 0.05 as significant. The arrows indicate the proteins of interest selected for identification and discussion
Differential expression of protein in different spots of interest selected on 2D gels of the roots of rice cultivars Rai Sudha and Munga Phool treated with low (T1, 10 mM) and high (T2, 25 mM) N doses. The values are the mean of three independent observations. Error bars indicate SE. Spot numbers are in accordance with 2D gels shown in Fig. 1
In Munga Phool, spots 2, 6, 9, 10 and 11 showed increased levels of expression at T1, but spots 1, 5 and 8 were under-expressed with reference to T0. A few spots, such as 3, 4 and 7, did not show any significant change. At T2 the increased level of spot expression continued in Rai Sudha, except for spots 2, 5, 6 and 10, which showed no significant increase in their intensity with reference to T1. In Munga Phool, the expression level decreased at T2 with reference to T1 for all the spots except 8, 9 and 11. Of the two genotypes, Rai Sudha showed a greater level of spot expression than Munga Phool at T1 (except for spot 9) and T2 (except for spots 8 and 9), in comparison with T0. The differential expression of the identified spots is discussed below in relation to various metabolic processes.
Energy metabolism
Glycolysis and gluconeogenesis
The metabolism of carbon- and nitrogen-containing compounds is fundamental to all forms of life. Spot 4 was similar to glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a tetrameric NAD-binding enzyme. Its expression was higher in LNE (Munga Phool) than in HNE (Rai Sudha) cultivar at T0 and T1. The condition was reversed at T2, where N treatments, in fact, enhanced the expression of this protein in Rai Sudha but reduced it in Munga Phool. Increase in the gene expression of GAPDH under increased nitrate supply has been reported earlier in Lycopersium esculentum roots (Wang et al. 2001; Wang et al. 2004) and Triticum aestivum grains (Flæte et al. 2005).
ATP synthesis
Spot 7 has been identified as a component of ATP synthase (alpha subunit), a key enzyme in oxidative phosphorylation in the photosynthesis pathway, located in chloroplasts and encoded by the chloroplast genome. It catalyzes synthesis of ATP from ADP during energy production in living beings (Wang et al. 2004). Its expression increased in Rai Sudha and decreased in Munga Phool in response to N at T2. This protein has also been identified in rice seedlings in response to different N levels (Kim et al. 2009), in drought-stressed maritime pine needles (Costa et al. 1998) and in low-oxygen-stressed maize roots (Huang et al. 2002). Nitrogen nutrition initiates the respiratory chain and enables ATP synthase to produce ATP for energy production (Lea and Ireland 1999).
Spot 6 was similar to cytochrome c, a small heme protein loosely associated with the inner membranes of mitochondria as a component of the electron-transport chain. It is also involved in the initiation of apoptosis as it binds to apoptotic protease-activating factor in cytoplasm. Expression of this protein spot increased in the HNE rice genotype with increase in N treatments. In the LNE genotype, its expression increased with T1 (10 mM N), but decreased with T2 (25 mM N).
Nitrogen metabolism
Spot 8 resembles glutamine synthetase (GS) enzyme, a key enzyme in nitrogen metabolism (Crawford 1995; Hakeem et al. 2011). In higher plants, inorganic N from the soil is converted into organic N by glutamine synthetase and glutamate synthase. GS catalyzes the ATP-dependent condensation of NH4 + with glutamate (Glu) to yield glutamine (Gln); GOGAT transfers the amide group of Gln to α-ketoglutarate (2-OG) to produce Glu (Cai et al. 2009). An individual N atom can pass through the GS reaction many times (Hirel and Lea 2001) during its uptake from the soil, assimilation and remobilization (Coque and Gallais 2006) prior to final deposition in a seed storage protein. In the root of Rai Sudha, the expression level of this spot increased significantly both at T1 and T2, while in Munga Phool it showed a 3-fold increase at T2 but a non-significant change at T1, with reference to T0. Studies on maize (Gallais et al. 2006) and wheat (Kichey et al. 2006) have suggested that GS activity is representative of the plant’s N status. Studies have shown that a substantial portion of N is not re-translocated to the harvested structures in the LNE cultivars and the GS activity in roots is lower than in the HNE cultivars (Fageria and Baligar 2005). The ability of the HNE cultivars to harvest more N and redistribute it to grains is due to their well-coordinated system of N uptake and assimilation. In LNE cultivars, GS activity was sufficiently high to maintain a low level of ammonium in leaf tissues irrespective of the external N supply (Habash et al. 2001); this explains the increase in activity of GS with increase in N supply.
Spot 11 has been identified as a porphobilinogen (PBG) deaminase. Its expression was upregulated in the HNE rice cultivar and down-regulated in the LNE cultivar with increase in N treatments. It catalyzes polymerization of four PBG monopyrrole units into the linear tetrapyrrole hydroxymethylbilane, leading to tetrapyrrole biosynthesis. The siroheme class of tetrapyrroles is a prosthetic group of nitrite and sulfite reductases, playing a central role in N and S assimilation, respectively. The role of siroheme in nitrite reductase is indicative of its significance for N assimilation in the biosphere; N assimilation increases the competition for ferrous ions under the inadequacy of siroheme. High expression of PBG deaminase in the HNE genotype under N treatments may be because this genotype contains adequate amounts of nitrate reductase and nitrite reductase required for efficient N assimilation.
Spot 2 of the root was identified as carbonic anhydrase (carbonate dehydratase), a family of zinc-containing enzymes that catalyze interconversion of CO2 and water into carbonic acid, protons and bicarbonate ions. Du Hyun et al. (2009) also identified this enzyme in response to low N supply to rice. Its expression increased significantly with N treatments in the HNE cultivar. However, in the LNE cultivar, the increase was significant at T1 only.
Sulfur metabolism
Spot 10 was similar to cysteine proteinase inhibitor-1 of the pyridoxal-phosphate-dependent enzyme family (EC 4.2.99.8). This enzyme has a role in cysteine biosynthesis, a key limiting step in the production of glutathione, which is a thiol implicated in resistance to environmental stresses. Its expression increased in the HNE genotype but declined significantly in the LNE genotype with increase in N treatments (both at T1 and T2). This shows an interaction between the cultivars and the N treatment levels, indicating the importance of N nutrition for sulfur assimilation (Grove et al. 2009). Constructive effects of N and S interaction on leaf area index, rate of photosynthesis and biomass production have also been observed (Qian and Schoenau 2007).
Putative disease-resistance response protein (spot 9) is among the defence-related proteins identified. Its induction under different N responses in roots suggests a potential link between N and plant vigour/plant ability to fight diseases. Du Hyun et al. (2009) identified a dirigent-like protein in response to N-treatment.
Oxidative defence system
Spot 5 corresponds to quinone oxidoreductase, a flavoprotein involved in electron-transport system. Generation of reactive oxygen species (ROS) by the mitochondrial electron-transport chain (ETC) is believed to be important in energy-generating processes. It might be involved in maintaining plant quinines in a reduced state in the cell. The quinone/quinol equilibrium may function in cells as a means of modulating the secondary metabolism, host–parasite interactions and growth-regulating mechanisms (Spitsberg and Coscia 2005). Quinone oxidoreductase protein protects cells against oxidative stress and toxic quinines. Increase in its expression with increase in N supply in the HNE genotype suggests its importance in sensing and counteracting the oxidative stress generated via the increased photorespiration and energy-generation mechanisms. However, its expression level decreased at both T1 and T2, with reference to T0.
Iron homeostasis
Spot 3 resembles an iron-storage protein (ferritin), possessing the eukaryotic ferritin domain. It has been shown that by introducing a soybean ferritin gene into rice, transgenic rice seeds stored three times more iron than the normal seeds (Goto et al. 1999; Qureshi et al. 2007). The expression level of this protein was lower at T0 in the HNE (Rai Sudha) than in the LNE (Munga Phool) cultivars. Surprisingly, it is one of the few proteins shown to decrease heavily under low N conditions in the HNE genotypes, suggesting its regulation by the availability of N in the system. It may therefore serve as one of the important marker proteins for selecting N-efficient rice cultivars.
Spot 1 is a putative myb-related protein, with a function in DNA binding. Its expression level increased at high N levels, but declined subsequently with further elevation of N.
From the results discussed above, the resemblances observed in spots 3, 8 and 10 were on the basis of the peptide mass fingerprinting account followed by a query match through Mascot search. The MW/pI ratio report confirms their resemblance. However, for any given protein spot resemblance, there may be several isoforms present or indeed other proteins, which needs to be confirmed in future studies.
Conclusions
The present study provides information for interpreting a physiology-based selection of N-related genotypes on the basis of proteomic results, with the aim of unravelling the molecular differences in rice root tissue grown under low, medium and high N supply. Comparative proteomics among genotypes for low N response is necessary for precise identification of potential key enzymes, which will lead to a better understanding of N-uptake and assimilation process in cereal crops. This will assist breeders in screening N-efficient cultivars and help in understanding how rice adapts to low N availability.
Abbreviations
- CHCA:
-
Cyanohydroxycinnamic acid
- DDW:
-
Double-distilled water
- DW:
-
Dry weight
- ETC:
-
Electron-transport chain
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
- γ-GHA:
-
γ-Glutamyl hydroxamate
- Gln:
-
Glutamine
- Glu:
-
Glutamate
- GOGAT:
-
Glutamate synthase
- GS:
-
Glutamine synthetase
- HNE:
-
High nitrogen efficient
- LNE:
-
Low nitrogen efficient
- MNE:
-
Moderate nitrogen efficient
- NE:
-
Nitrogen efficiency
- NiR:
-
Nitrite reductase
- NR:
-
Nitrate reductase
- NUE:
-
Nitrogen use efficiency
- PBG:
-
Porphobilinogen
- PMF:
-
Peptide mass fingerprinting
- ROS:
-
Reactive oxygen species
References
Abrol YP, Chatterjee SR, Kumar PA, Jain V (1999) Improvement in nitrogenous fertilizer utilization—physiological and molecular approaches. Curr Sci 76:1357–1364
Agarwal GK, Rakhwal R, Yonekura M, Kubo A, Saji H (2002) Proteome analysis of differentially displayed proteins as a tool for investigating ozone stress in rice (Oryza sativa L.) seedlings. Proteomics 2:947–959
Ahmad A, Abdin MZ (1999) NADH: nitrate reductase and NAD (P) H: nitrate reductase activity in mustard seedlings. Plant Sci 143:1–8
Ahmad A, Khan I, Anjum NA, Diva I, Abrol YP, Iqbal M (2005) Effect of timing of sulphur fertilizer application on growth and yield of rapeseed (Brassica campestris L.). J Plant Nutr 28:1049–1059
Ahmad A, Khan I, Abrol YP, Iqbal M (2008) Genotypic variation of nitrogen use efficiency in Indian mustard. Environ Pollut 154:462–466
Anjana UmarS, Iqbal M (2007) Nitrate accumulation in plants, factors affecting the process, and human health implications: a review. Agron Sustain Dev 27:45–57
Aslam M, Travis RL, Huffaker RC (1992) Comparative kinetics and reciprocal inhibition of nitrate and nitrite uptake in roots of uninduced and induced barley (Hordeum vulgare L.) seedlings. Plant Physiol 99:1124–1133
Bahrman N, Gouy A, Devienne-Barret F, Hirel B, Vedele F, Le Gouis J (2005) Differential change in root protein patterns of two wheat varieties under high and low nitrogen nutrition levels. Plant Sci 168:81–87
Bloom AJ (1985) Wild and cultivated barleys show similar affinities for mineral nitrogen. Oecologia 65:555–557
Bradford M (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254
Cai H, Zhou Y, Xiao J, Li X, Zhang Q, Lian X (2009) Overexpressed glutamine synthetase gene modifies nitrogen metabolism and abiotic stress responses in rice. Plant Cell Rep 28:527–537
Cao Y, Fan XR, Sun S, Xu G, Hu J, Shen Q (2008) Effect of nitrate on activities and transcript levels of nitrate reductase and glutamine synthetase in rice. Pedosphere 18:664–673
Carezo M, Garcia-Agustin P, Serna MD, Primo-Millo E (1997) Kinetics of nitrate uptake by citrus seedlings and inhibitory effects of salinity. Plant Sci 126:105–112
Chandna R, Gupta S, Ahmad A, Iqbal M, Prasad M (2010) Variabilities in Indian bread wheat (Triticum aestivum L.) varieties differing in nitrogen efficiency as assessed by microsatellite markers. Protoplasma 242:55–67
Clarkson DT (1985) Factors affecting mineral nutrient acquisition by plants. Annu Rev Plant Physiol 36:77–115
Coque M, Gallais A (2006) Genomic regions involved in response to grain yield selection at high and low nitrogen fertilization in maize. Theor App Genet 112:1205–1220
Conn SJ, Hocking B, Dayod M, Xu B, Athman A, Henderson S, Aukett L, Conn V, Shearer MK, Fuentes S, Tyerman SD, Gilliham M (2013) Protocol: optimising hydroponic growth systems for nutritional and physiological analysis of Arabidopsis thaliana and other plants. Plant Methods 9:4
Costa P, Bahrman N, Frigerio JM, Kermer A, Plomion C (1998) Compression wood-responsive proteins in developing xylem of maritime pine (Pinus pinaster Ait.). Plant Mol Biol 38:587–596
Crawford NM (1995) Nitrate: nutrient and signal for plant growth. Plant Cell 7:859–868
Du Hyun K, Junko S, Dea-Wook K, Myung Kyu O, Myung Ki K, Sung SI, Hitoshi I, Yoshinori M, Randeep R (2009) Gel-based proteomics approach for detecting low nitrogen-responsive proteins in cultivated rice species. Physiol Mol Biol Plants 15:31–41
Evans HJ, Nason A (1953) Pyridine nucleotide-nitrate reductase from extracts of higher plants. Plant Physiol 28:233–254
Fageria NK, Baligar VC (2005) Enhancing nitrogen use efficiency in crop plants. Adv Agron 88:97–185
Flæte NES, Hollung K, Ruud L, Sogn T, Færgestad EM, Skarpeid HJ, Magnus EM, Uhlen AK (2005) Combined nitrogen and sulphur fertilisation and its effect on wheat quality and protein composition measured by SE-FPLC and proteomics. J Cer Sci 41:357–369
Gallais A, Coque M, Quilleré I, Prioul JL, Hirel B (2006) Modelling post-silking N-fluxes in maize using 15N-labelling field experiments. New Phytol 172:696–707
Goto F, Yoshihara T, Shigemoto N, Toki S, Takaiwa F (1999) Iron fortification of rice seed by the soybean ferritin gene. Nat Biotechnol 17:282–286
Goyal SS, Huffaker RC (1986) The uptake of NO3– and NH4 + by intact wheat (Triticum aestivum) seedlings. Plant Physiol 82:1051–1056
Grove H, Hollung K, Moldestad A, Færgestad EM, Uhlen AK (2009) Proteome changes in wheat subjected to different nitrogen and sulfur fertilizations. Agric Food Chem 57:4250–4258
Habash DZ, Massiah AJ, Rong HL, Wallsgrove RM, Leigh RA (2001) The role of cytosolic glutamine synthetase in wheat. Ann Appl Biol 138:83–89
Hakeem KR, Ahmad A, Iqbal M, Gucel S, Ozturk M (2011) Nitrogen-efficient cultivars can reduce nitrate pollution. Environ Sci Pollut Res 18:1184–1193
Hakeem KR, Khan F, Chandna R, Siddiqui TO, Iqbal M (2012a) Genotypic variability among soybean genotypes under NaCl stress and proteome analysis of salt tolerant genotype. App Biochem Biotech 168:2309–2329
Hakeem KR, Chandna R, Ahmad A, Iqbal M (2012b) Physiological and molecular analysis of applied nitrogen in rice (Oryza sativa L.) genotypes. Rice Sci 19(3):213–222
Hirel B, Lea PJ (2001) Ammonia assimilation. In: Lea PJ, Morat-Gaudry JF (eds) Plant nitrogen. Springer, Berlin, pp 79–99
Hirel B, Tercé-Laforgue MartinT, Gonzalez-Moro MB, Estavillo JM (2005) Physiology of maize. I. A comprehensive and integrated view of nitrogen metabolism in a C4 plant. Physiol Plant 124:167–177
Huang XQ, Börner A, Röder MS, Ganal MW (2002) Assessing genetic diversity of wheat (Triticum aestivum L.) germplasm using microsatellite markers. Theor Appl Genet 105:699–707
Hunt J, Seymour DJ (1985) Method for measuring nitrate-nitrogen in vegetables using anion-exchange high performance liquid chromatography. Analyst 110:131–133
Khan F, Hakeem KR, Sidiqqui TO (2013) RAPD markers associated with salt tolerance in Soybean genotypes under salt stress. App Biochem Biotech 170(2):257–272
Kichey T, Heumez E, Pocholle P, Pageau K, Vanacker H, Dubois F, Le Gouis J, Hirel B (2006) Combined agronomic and physiological aspects of nitrogen management in wheat (Triticum aestivum L.). Dynamic and integrated views highlighting the central role for the enzyme glutamine synthetase. New Phytol 169:265–278
Kim ST, Kim SG, Hwang DH, Kang SY (2004) Proteomic analysis of pathogen-responsive proteins from rice leaves induced by rice blast fungus, Magnaporthe grisea. Proteomics 4:3569–3578
Kim SR, Yang J, Moon S, Ryu CH, An K, Kim KM, Yim J, An G (2009) An rice OGR1 encodes a pentatricopeptide repeat—DYW protein and is essential for RNA editing in mitochondria. Plant J 59:738–749
Lea PJ, Ireland RJ (1999) Nitrogen metabolism in higher plants. In: Singh BK (ed) Plant amino acids. Marcel Dekker Inc., New York, pp 1–47
Masclaux C, Quillere I, Gallis A, Hirel B (2001) The challenge of remobilization in plant nitrogen economy. A survey of physio-agronomic and molecular approaches. Ann Appl Biol 138:69–81
McNally SF, Hirel B, Gadal P, Mann AF, Stewart GR (1983) Glutamine synthethase of higher plants: evidences for a specific isoform content related to their possible physiological role and their compartmentation within the leaf. Plant Physiol 72:22–25
Natarajan S, Xu C, Caperna TJ, Garrett WM (2005) Comparison of protein solubilization methods suitable for proteomic analysis of soybean seed proteins. Anal Biochem 342:214–220
Newsholme SJ, Maleeft BF, Steiner S, Anderson NL, Schwartz LW (2000) Two-dimensional electrophoresis of liver proteins: characterization of a drug-induced hepatomegaly in rats. Electrophoresis 2:2122–2128
Neyra A, Hageman RH (1976) Relationships between carbon dioxide, malate and nitrate accumulation and reduction in corn (Zea mays L.) seedlings. Plant Physiol 58:726–730
Pace GM, McClure PR (1986) Comparison of nitrate uptake parameters across maize inbred lines. J Plant Nutr 9:1095–1111
Pathak RR, Ahmad A, Lochab S, Raghuram N (2008) Molecular physiology of plant nitrogen use efficiency and biotechnological options for its enhancement. Curr Sci 94:1394–1403
Peng Z, Wang M, Li F, Lv H, Li C, Xia G (2009) A proteomic study of the response to salinity and drought stress in an introgression strain of bread wheat. Mol Cell Proteomics 8(12):2676–2686
Perkins DN, Pappin DJC, Creasy DM, Cottrell JS (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20:3551–3567
Qian P, Schoenau JJ (2007) Using an anion exchange membrane to predict soil available N and S supplies and the impact of N and S fertilization on canola and wheat growth. Pedosphere 17:77–83
Qureshi MI, Qadir S, Zolla L (2007) Proteomics-based dissection of stress-responsive pathways in plants. J Plant Physiol 164:1239–1260
Qureshi MI, D’Amici GM, Fagioni M, Rinalducci S, Zolla L (2010) Iron stabilizes thylakoid protein-pigment complexes in Indian mustard grown under Cd-stress as revealed by BN-SDS-PAGE and ESI-MS/MS. J Plant Physiol 167:761–770
Rhodes D, Sims AP, Stewart GR (1975) The control of glutamine synthetase levels in Lemna minor. Planta 125:201–211
Smil V (1999) Nitrogen in crop production: an account of global flows. Global Biogeochem Cycles 13:647–662
Smith IK (1980) Regulation of sulfate assimilation in tobacco cells: effect of nitrogen and sulphur nutrition on sulfate permease and O-acetylserine sulfhydrylase. Plant Physiol 66:877–883
Song X, Ni Z, Yao Y, Xie C, Li Z, Wu H, Zhang Y, Sun Q (2007) Wheat (Triticum aestivum L.) root proteome and differentially expressed root proteins between hybrid and parents. Proteomics 7:3538–3557
Spitsberg VL, Coscia CJ (2005) Quinone reductases of higher plants. Eur J Biochem 127:67–70
Wang YH, Garvin DF, Kochian LV (2001) Nitrate-induced genes in tomato roots. Array analysis reveals novel genes that may play a role in nitrogen nutrition. Plant Physiol 127:345–359
Wang X, Bian Y, Cheng K, Zou H, Sai-Ming Sun S, He J (2012) A comprehensive differential proteomic study of nitrate deprivation in arabidopsis reveals complex regulatory networks of plant Nitrogen Responses. J Proteome Res 11(4):2301–2315
Wang SB, Chen F, Sommerfeld M, Hu Q (2004) Proteomic analysis of molecular response to oxidative stress by the green alga Haematococcus pluvialis (Chlorophyceae). Planta 220:17–29
Woodend JJ, Glass AMD, Person C (1986) Intraspecific variation for nitrate uptake and nitrogen utilization in wheat (T. aestivum L.) grown under nitrogen stress. J Plant Nutr 9:1213–1225
Acknowledgments
K.R.H. is grateful to the University Grants Commission, Government of India, for granting him a Junior Research Fellowship. M.I.Q. is grateful to DST and UGC, Govt. of India. Part of the study was done when M.I. was Visiting Professor at the Plant Production Department of King Saud University, Riyadh, Saudi Arabia.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Hakeem, K.R., Mir, B.A., Qureshi, M.I. et al. Physiological studies and proteomic analysis for differentially expressed proteins and their possible role in the root of N-efficient rice (Oryza sativa L.). Mol Breeding 32, 785–798 (2013). https://doi.org/10.1007/s11032-013-9906-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-013-9906-0