Abstract
Powdery mildew limits cucumber production worldwide. Most resistant cucumber cultivars become susceptible to powdery mildew at low temperatures. Resistance within a wide temperature range is therefore desirable for cucumber production. We constructed a cucumber genetic linkage map based on a population of 111 recombinant inbred lines derived from a cross between CS-PMR1, with strong and temperature-independent resistance, and Santou, with moderate and temperature-dependent resistance. The map spans 693.0 cM and consists of 296 markers segregating into seven linkage groups; the markers include 289 simple sequence repeats (SSRs), six sequence characterized amplified regions, and one inter simple sequence repeat. Due to the presence of 150 common SSR markers, we were able to compare our map with previously published maps obtained by using populations derived from inter- or intra-variety crosses. We also evaluated powdery mildew resistance of the recombinant inbred lines and identified seven quantitative trait loci (QTL) contributed by CS-PMR1 and two QTL contributed by Santou. Four QTL (pm3.1, pm5.1, pm5.2 and pm5.3) were successfully validated by using populations derived from residual heterozygous lines. Some of the QTL identified in our study are in good agreement with previously published results obtained with materials of different origin. The markers reported here would be useful for introducing high and temperature-independent resistance by accumulation of QTL from CS-PMR1 and Santou.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Powdery mildew is a severe disease of cucumber (Cucumis sativus L.) caused by Podosphaera xanthii (syn. Sphaerotheca fuliginea ex Fr. Poll.) and, to a lesser degree, Golovinomyces cichoracearum (syn. Erysiphe cichoracearum DC ex Mérat.). At present, fungicide application is the major method of disease control, but it is difficult to apply fungicides at the time of disease outbreak, since cucumber fruits are harvested daily. In addition, the labor and cost of fungicide application impose heavy burdens on farmers. Thus, breeding for powdery mildew resistance is the most desirable approach for disease control. Although a large number of resistant accessions from South and East Asia have been identified and utilized to develop new resistant varieties, resistance of cucumber varieties remains insufficient (Morishita et al. 2003). Morishita et al. (2003) observed that most cucumber varieties and accessions are resistant to powdery mildew at 26 °C, but susceptible at 20 °C. Among 295 cucumber accessions tested, PI 197088-5 displayed the highest level of resistance both at 26 and 20 °C; its resistance was suggested to be controlled by two genes, one recessive and the other incompletely dominant.
The availability of genomic information, such as whole genome sequences, expressed sequence tag sequences, and high-density genetic linkage maps, facilitates transition from traditional to molecular breeding. Among Cucurbitaceae (which include such species as cucumber, melon, watermelon, and squash), molecular breeding of melon is at the most advanced stage. A melon consensus linkage map has been published (Diaz et al. 2011), composed of 1,592 markers, including markers for 11 phenotypic traits, and 370 quantitative trait loci (QTL) controlling 62 traits (such as resistance to powdery mildew and to cucumber mosaic virus). As the consensus map has been constructed primarily by using highly transferable anchor markers, it will be useful for increasing the effectiveness and efficiency of breeding via marker-assisted selection. Also published recently are a cucumber genome sequence (Huang et al. 2009), genetic linkage maps constructed based on transferable markers (Fukino et al. 2008b; Ren et al. 2009; Miao et al. 2011), and an integrated genetic map (Zhang et al. 2012); however, there is not enough information on DNA markers that can be used for marker-assisted selection in cucumber. Several linkage maps have been published that include phenotypic traits, such as resistance to zucchini yellow mosaic virus (Park et al. 2004) and to downy mildew (Horejsi et al. 2000), or QTL, such as powdery mildew resistance (Sakata et al. 2006; Liu et al. 2008a, b). Likewise, two or three QTL for powdery mildew resistance of PI 197088-1 (a sib-line of PI 197088-5) have been detected, and sequence characterized amplified region (SCAR) markers associated with each QTL have been developed (Sakata et al. 2006). However, since these maps were constructed based mainly on dominant markers, they lack versatility and do not apply to all populations. There are few reports on universal markers linked to agronomically useful traits (Weng et al. 2010; Miao et al. 2011; Kang et al. 2011).
The objective of this study was to conduct a QTL analysis of powdery mildew resistance of recombinant inbred lines (RILs) derived from a cross between CS-PMR1 [previously referred to as PI 197088-1 by Sakata et al. (2006) and Fukino et al. (2008b)] and Santou varieties. For accurate analysis, resistance was evaluated at two developmental stages. In addition, the QTL effectiveness was confirmed by using residual heterozygous lines (RHLs), i.e. lines chosen from RILs that have heterozygous segments surrounding the powdery mildew resistance QTL (Yamanaka et al. 2005).
Materials and methods
Plant materials and genomic DNA isolation
The population of 111 F8 RILs was derived by single seed descent from a cross between CS-PMR1 (a weedy cucumber of Indian origin, highly resistant to powdery mildew in a temperature-independent manner) and Santou (a Japanese native cucumber cultivar, moderately resistant to powdery mildew) (Sakata et al. 2006). Although these RILs were derived from the same cross as used in Sakata et al. (2006) and Fukino et al. (2008b), the size of the population varied slightly. Genomic DNA was isolated from young leaves of the parents and their RILs with a DNeasy 96 Plant Kit (Qiagen, Tokyo, Japan), and used for marker analysis.
Powdery mildew resistance tests
Out of 111 RILs, 109 were used for powdery mildew resistance tests (two RILs failed to produce enough seeds). The two parental lines were also included.
Leaf disc assay
Resistance to powdery mildew was tested by using the pxA strain of P. xanthii (Fukino et al. 2008a). Plants were sown in a greenhouse (three plants per line). At the second leaf stage, two leaf discs (15 mm in diameter) per plant were cut from the first leaves of two plants and placed (one disc per plant) on each of two similar square dishes containing 0.8 % agar (224 × 224 mm; 400 ml). Inoculation was performed by spraying with a conidial suspension according to Morishita et al. (2003). One dish was incubated at 25 °C (experiment LD25) and the other at 20 °C (LD20), both under a 16:8-h light:dark photoperiod. After 2 weeks, the degree of sporulation was scored visually according to Fukino et al. (2004) on a scale of 0 (no sporulation) to 5 (heavy sporulation on the entire disc), and used as a disease index (DI). Experiments were replicated five times. The average DI from five independent tests for each line was used in QTL analysis.
Evaluation of naturally occurring disease in plastic greenhouses
Experiments were conducted in the late spring to summer (experiment GH-S) and in the autumn (GH-A) of 2009 at the NARO Institute of Vegetable and Tea Science, Tsu, Mie, Japan. Both GH-S and GH-A experiments used a completely randomized design with two replications (one plant per line per replication). Seeds were sown in a greenhouse on May 7 and Sept. 10, and seedlings were transplanted to individual plastic houses on May 21 and Sept. 29, respectively. Disease caused by naturally occurring powdery mildew was assessed on July 22 (GH-S) and Dec. 4 (GH-A) as described above for the leaf disc assay. The average outside temperature was determined for 30 days before each analysis: it was 25.0 and 13.9 °C, respectively. In the autumn, the plastic houses were warmed by a heater to maintain the temperature above 15 °C, which is required for cucumber growth. The average DI from the two replications was used for QTL analysis.
Marker analysis
We previously developed an SSR-based genetic linkage map, which comprised 120 simple sequence repeats (SSRs) and six SCARs (Fukino et al. 2008b). Two markers (CS08 and CS39) from previous map were discarded because of poor reproducibility. One hundred and seventy-one SSR markers from melon and cucumber were added: three CSAT/CSCT/CMCCA (Danin-Poleg et al. 2001), one TJ (Gonzalo et al. 2005), one CS (Kong et al. 2006), five CM (Kong et al. 2007), six CMN (Fukino et al. 2007), two CSN (Fukino et al. 2008b), one CSJCT (Watcharawongpaiboon and Chunwongse 2008), 10 ECM/GCM (Fernandez-Silva et al. 2008), 141 SSR (Ren et al. 2009), and one CU (Cucurbit Genomics Database, http://www.icugi.org/). The markers were screened for polymorphisms between the parents and used for the construction of a genetic linkage map. SCAR markers (Sakata et al. 2006) and an inter simple sequence repeat (ISSR) marker (Alfandi et al. 2009) were also used, since they were previously reported to be linked to powdery mildew resistance.
Marker analyses were conducted as previously described (Fukino et al. 2008a) with minor modifications. For SSR analysis, PCR products were labeled by post-PCR labeling (Kukita and Hayashi 2002) by using R110-ddUTP (PerkinElmer, Yokohama, Japan). The sizes of the amplified fragments were estimated by using an automated DNA analyzer (3730xl or ABI PRISM 3100, Applied Biosystems, Tokyo, Japan). For ISSR analysis, polymorphism was detected by separating the amplified DNA on a 3.5 % Agarose XP (Nippon Gene, Tokyo, Japan) gel in TBE (Tris–borate–EDTA) buffer containing ethidium bromide. DNA bands were visualized under a UV transilluminator.
Linkage map construction and QTL detection
Standard χ 2 analysis was used to test the segregation pattern at each marker locus for deviations from the expected Mendelian ratio of 1:1 in RILs. The linkage map for the CS-PMR1 × Santou RIL population was constructed with MAPMAKER/EXP 3.0 (Lander et al. 1987). Marker data were assigned to linkage groups (LGs) by using a minimum logarithm of odds (LOD) likelihood score of 5.0. The Kosambi map function (Kosambi 1943) was used to calculate the genetic distance between markers. The previously reported assignment of LGs to corresponding chromosomes (Ren et al. 2009) was used. QTL analysis was performed by using both composite interval mapping (Zeng 1993, 1994) with Windows QTL Cartographer v.2.5 (Wang et al. 2007) and the Kruskal–Wallis nonparametric rank-sum test (Lehmann 1975) with MapQTL (van Ooijen 2004). Our new data and the data from the experiments by Sakata et al. (2006) (S26 and S20, performed at 26 or 20 °C, respectively) were used to estimate putative QTL from the calculated LOD threshold scores after 1,000 permutation tests (≥3.0 LOD for LD20; ≥2.8 LOD for LD25, GH-A, S26 and S20; ≥2.7 LOD for GH-S).
QTL validation by using RHLs
Six populations derived from selfing of six RHLs were used for QTL validation. Markers that mapped near the QTL were used for genotyping. Sixty-nine to 104 plants derived from selfing of each RHL were used for marker genotype identification and assessment of powdery mildew resistance. Genomic DNA was isolated from cotyledons of the individual plants on 0.2-μm polyvinylidene difluoride filter plates and glass fiber filter plates (Corning #3504 and #3511, respectively; Corning, Tokyo, Japan) as described by Fukino et al. (2012). Powdery mildew resistance of these populations was tested as described under ‘Leaf disc assay’ with minor modifications. Four discs per plant were cut and the test was conducted once (with two replications) for each population. Plants within a population were classified according to the marker genotypes; recombinants between the two markers and plants with heterozygous marker genotypes were excluded. The mean DI values were calculated for each class. Differences between the means were statistically analyzed by using the Mann–Whitney U-test, with significance at P < 0.05.
Results
Evaluation of powdery mildew resistance
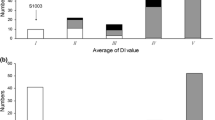
CS-PMR1 was completely resistant in all experiments, whereas Santou was moderately resistant (LD25 and GH-S) to susceptible (LD20 and GH-A) (Fig. 1). Average DI values for each RIL were continuously distributed from resistant to susceptible, suggesting polygenic control of this trait. Greater resistance was observed in RILs in the LD25 and GH-S experiments than in LD20 and GH-A. Plants with higher DI scores (more susceptible) than Santou were found in all experiments, which indicates transgressive segregation.
Frequency distribution of average disease index (DI) scores in RILs derived from Cucumis sativus CS-PMR1 × Santou assessed by the leaf disc assay (a) and by evaluation of naturally occurring disease caused by Podosphaera xanthii (b). I DI = 0; II 0 < DI ≤ 1; III 1 < DI ≤ 2; IV 2 < DI ≤ 3; V 3 < DI ≤ 4; VI 4 < DI ≤ 5
Construction of a linkage map
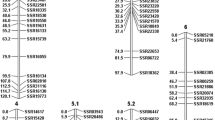
The CS map was constructed based on 296 markers (289 SSRs, six SCARs, and one ISSR) (Fig. 2). The resulting map contains seven LGs spanning 693.0 cM, with an average distance between markers of 2.3 cM. Linkage groups were previously assigned to chromosomes by Ren et al. (2009). Of 296 mapped markers, 74 (25 %) deviated from the expected 1:1 segregation ratio (P < 0.05): 14, 27, 13, 1, 14 and 5 deviating markers were found on chromosomes 1, 2, 3, 4, 5 and 6, respectively. There were no deviating markers on chromosome 7. Among five SCARs reported to be linked to the QTL for powdery mildew resistance (Sakata et al. 2006), EAAGMCAT280-282STS, EAACMCAC391-395STS and EAAGMCAG154STS were mapped to chromosome 1; EAACMCTG116STS to chromosome 5; and EAAGMCTG171-179STS to chromosome 7. An ISSR marker UBC809 was mapped to chromosome 5, located 11.8 cM from EAACMCTG116STS.
Linkage map for C. sativus CS-PMR1 × Santou RILs. The genotypes of 111 RILs were determined by using 289 SSR, six SCAR, and one ISSR markers. The markers are shown to the right of the linkage groups (1–7), and recombination distances are indicated in centimorgans to the left. Linkage groups were assigned to chromosomes by Ren et al. (2009). Underlined markers deviated from the expected segregation ratio. The markers are denoted using the following nomenclature: CM Kong et al. (2007), CMNC10 Fukino et al. (2007), ECM Fernandez-Silva et al. (2008), SSR Ren et al. (2009). The sources of all other markers have been described previously (Fukino et al. 2008b)
We compared our CS map with the hs (Ren et al. 2009) and ss (Miao et al. 2011) maps (ESM 1 and ESM 2); this comparison was made possible by the presence of 150 and 30 common markers, respectively. The major difference between the CS and hs maps was the total length of chromosomes 4 and 5, which were 65 and 41 % shorter on the hs map. A discrepancy in the marker order on these chromosomes between the CS and hs maps was also observed. The CS and ss maps were similar, except for the length of chromosome 4 (106.3 vs. 50.5 cM, respectively).
QTL analysis
Six, four, three and four significant QTL were detected by composite interval mapping in the experiments LD25, LD20, GH-S and GH-A, respectively (Table 1). Each QTL accounted for 0.05–0.24 of the variance of the trait. QTL pm1.2 and pm 6.1 were detected in all four experiments, whereas pm4.1 and pm5.3 were common to LD25, LD20 and GH-A; pm2.1 and pm3.1 were found only in LD25; and pm5.2 was detected only in GH-S. No temperature-dependent QTL were reproducibly detected. Santou contributed resistance to powdery mildew at pm5.2 and pm6.1, whereas CS-PMR1 contributed resistance at the other QTL. The LOD peak positions for the QTL pm4.1 were slightly different in different experiments: 74.2 and 87.9 cM in LD25, 92.9 cM in LD20, and 74.2 cM in GH-A (ESM 3). The largest QTL also differed depending on the experiment (pm6.1 in LD25, pm5.3 in LD20, pm5.2 in GH-S, and pm1.2 in GH-A).
Re-analysis of the data of Sakata et al. (2006) resulted in detection of four QTL (pm1.1, pm5.2, pm5.3 and pm6.1) for S26 and three QTL (pm1.1, pm5.1 and pm5.2) for S20 (Table 1). QTL pm1.1 and pm5.1, contributed by CS-PMR1, were novel, whereas QTL pm5.2, pm5.3 and pm6.1 were confirmed by our analysis. The nonparametric Kruskal–Wallis rank-sum test showed highly significant (P < 0.01) association of the markers mapped on the regions surrounding detected QTL.
To compare the QTL present in resistant and in susceptible RILs, the DI cut-off values were chosen so that the top ca. 10 % and the bottom ca. 10 % of RILs were selected. The resistant group comprised 14 RILs with DI ≤ 1 in all experiments; the susceptible group was represented by 11 RILs with DI ≥ 3.5 in LD20 and GH-A (Table 2). Among nine QTLs, resistant RILs had more than three, and susceptible RILs had less than five. More than 75 % of resistant RILs had QTL pm1.1, pm1.2, pm2.1, pm5.1 and pm6.1, whereas less than 30 % of susceptible RILs had pm1.1, pm1.2, pm5.1, pm5.3 and pm6.1.
QTL validation by using RHLs
Populations derived from selfing of six RHLs were evaluated for powdery mildew resistance and genotyped with the markers corresponding to the heterozygous regions of RHLs (Table 3). Significant differences between two marker genotypes were found at both 25 and 20 °C in populations 1, 5 and 6 (QTL pm3.1, pm5.2 and pm5.3, respectively; Table 3). In population 4, which has a heterozygous region surrounding QTL pm5.1, significant difference was observed only at 25 °C. As pm5.2 is the QTL contributed by Santou, the average DI score of the plants with Santou genotype was lower than that of the CS-PMR1 plants in population 5. The average DI scores of CS-PMR1 plants in populations 1 and 6 were lower than those of Santou plants. Population 2, which had a heterozygous region surrounding QTL pm3.1, was completely resistant in both experiments regardless of the marker genotype. In population 3, which had a heterozygous region surrounding QTL pm4.1, plants were highly resistant at 25 °C, but moderately resistant at 20 °C regardless of their genotypes, with no significant differences among the average DI scores. This might be because the effect of QTL pm4.1 was not fully expressed in the genetic background of population 3, or because the position of this QTL was beyond the heterozygous region.
Discussion
Cucumber linkage map
A quarter of the mapped markers deviated from the expected segregation ratio, with the largest clusters of markers with distorted segregation (27, 13 and 14) found on chromosomes 2, 3 and 5, respectively. Segregation distortion and marker clustering are often reported in cucumber (Yuan et al. 2008; Weng et al. 2010; Miao et al. 2011), and 29.5, 6.3 and 18.0 % of markers with distorted segregation were observed in the respective studies. The reason for this phenomenon has not yet been clarified. Differences in the marker order and distances were observed between the maps derived from the inter-varietal (hs map) and the intra-varietal (CS and ss maps) crosses, possibly due to chromosome rearrangements between C. sativus var. sativus and C. sativus var. hardwickii (Ren et al. 2009). In particular, chromosomes 4 and 5 were much shorter on the hs map than on the CS map, and discrepancies in the marker order were also detected on these chromosomes. The latter results agree with the observations by Miao et al. (2011), who reported discrepancies between the ss and hs maps in the marker order on chromosomes 4, 5 and 7. No discrepancies in chromosome 7 length and marker order were observed between the CS and hs maps; this might be due to the relatively small number of markers (18) on chromosome 7 on the CS map. The markers on chromosomes 1 and 4 reported on the ss map (17 and 15, respectively) seem to be under-represented, as their numbers are less than half of those on the CS map (43 and 33, respectively).
Our CS map, together with the ss map, will complement the hs map. As the CS and hs maps have 150 markers in common, fine mapping of the regions of interest will be easier by using the hs map as a reference.
QTL for powdery mildew resistance
In agreement with Morishita et al. (2003), a high level of temperature-independent resistance was observed in CS-PMR1 in all experiments, and a trend towards a higher resistance at high temperature was observed in RILs. A gene conferring resistance to wheat stripe rust at high (but not at low) temperature has been found in wheat (Fu et al. 2009). The 20 °C-specific QTL from CS-PMR1 on LG III (chromosome 7) and from Santou on LG IV (chromosome 5) were reported in cucumber (Sakata et al. 2006), which suggested that a temperature-dependent gene may confer resistance in each of these cultivars. However, no QTL was detected on chromosome 7 in the current study. The QTL on LG IV corresponds to our pm5.2 detected in GH-S, S26 and S20, and thus cannot be considered as a temperature-specific QTL. A novel QTL pm5.1 was detected in S20 by our re-analysis of Sakata et al.’s (2006) data, but not in any experiments in our current study. By using a population derived from RHLs, we detected a significant effect of pm5.1 at 25 °C but not at 20 °C. Therefore, pm5.1 also appears not to be temperature-specific. Interestingly, population 3, which has a heterozygous region around QTL pm4.1, showed obvious temperature-dependent resistance unrelated to the genotype of pm4.1. This population could be an essential tool for the analysis of temperature-dependent resistance. The effect of some QTL may be weak and undetectable under some conditions, so in spite of common parental lines used for the cross, the results of our current QTL analysis and our re-analysis data of Sakata et al. (2006) were not identical. The QTL also slightly varied between different experiments, probably due to different developmental stages and assay methods (leaf discs of plantlets vs. greenhouse evaluation of adult plants).
QTL for powdery mildew resistance were previously reported by using populations derived from a cross between a resistant breeding line S06 (European greenhouse type) and a susceptible inbred line S94 (Northern China open-field type) (Liu et al. 2008a, b). It is difficult to directly compare the QTL positions determined in our and the above studies, as most of the markers used by Liu et al. (2008a,b) were sequence-related amplified polymorphism markers, which are absent on our linkage map. Potentially, markers surrounding QTL for powdery mildew resistance (Liu et al. 2008a, b) could be assigned to our linkage map by using common markers in a high-density integrated genetic map (Zhang et al. 2012). The positions of PM1.2 (Liu et al. 2008b), pm6.1 (Liu et al. 2008a) and PM2.2 (Liu et al. 2008b) appear to be close to the positions of pm1.2, pm5.1 and pm6.1 in our study, respectively. Furthermore, an ISSR marker UBC809 reported to be linked to powdery mildew resistance of the cucumber accession JIN5 (Alfandi et al. 2009) was mapped on chromosome 5, adjacent to pm5.2 contributed by Santou. Concentration of the resistance genes in certain genomic regions has been suggested for rice blast (Fukuoka and Okuno 2001) and melon powdery mildew (Fukino et al. 2008a). This might also be the case with cucumber, as four QTL (pm1.2, pm5.1, pm5.2 and pm6.1) detected in our study might be located in the vicinity of QTL detected previously by using different resistant materials. The markers linked to the QTL detected in our study might be useful for selection of powdery mildew resistance in different cucumber accessions.
The QTL detected varied depending on the tests, but the effects of four of them (pm3.1, pm5.1, pm5.2 and pm5.3) were successfully confirmed by using populations derived from RHLs. As there were no RHLs corresponding to all QTL, the effects of other QTL are yet to be confirmed. Although CS-PMR1 is highly resistant to powdery mildew, it has rarely been used for breeding, since it looks quite different from cultivated cucumbers and has poor fruit characteristics (Sakata et al. 2008). Thus, many backcrosses would be needed to breed commercial cultivars if CS-PMR1 were to be used, and it would be difficult to maintain resistance in the progeny due to its polygenic inheritance. It may be necessary to accumulate three or more QTL in order to breed for strong and temperature-independent resistance. Because most of the resistant RILs had them, pm1.1, pm1.2, pm2.1, pm5.1 and pm6.1 would be good candidate QTL for this purpose. Interestingly, pm6.1, present in all resistant RILs, was contributed by the moderately resistant parent Santou. Combination of QTL from CS-PMR1 and Santou might result in higher resistance by accumulation of fewer resistance genes. Marker-assisted selection will provide an effective tool for breeding cucumbers highly resistant to powdery mildew. The markers linked to QTL will be useful for introducing powdery mildew resistance genes of CS-PMR1, Santou, or both into commercial cultivars to enhance resistance. Map-based cloning and subsequent analysis of gene functions may help to determine the key players in the high and temperature-independent cucumber resistance to powdery mildew.
References
Alfandi M, Shen L, Qi X, Xu Q, Chen X (2009) A SCAR marker linked to powdery mildew resistance for selection of the near-isogenic lines in cucumber. In: Abstracts of the 4th international Cucurbitaceae symposium, pp 20–21
Danin-Poleg Y, Reis N, Tzuri G, Katzir N (2001) Development and characterization of microsatellite markers in Cucumis. Theor Appl Genet 102:61–72
Diaz A, Fergany M, Formisano G et al (2011) A consensus linkage map for molecular markers and quantitative trait loci associated with economically important traits in melon (Cucumis melo L.). BMC Plant Biol 11:111. doi:10.1186/1471-2229-11-111
Fernandez-Silva I, Eduardo I, Blanca J, Esteras C, Picó B, Nuez F, Arús P, Garcia-Mas J, Monforte AJ (2008) Bin mapping of genomic and EST-derived SSRs in melon (Cucumis melo L.). Theor Appl Genet 118:139–150. doi:10.1007/s00122-008-0883-3
Fu D, Uauy C, Distelfeld A, Blechl A, Epstein L, Chen X, Sela H, Fahima T, Dubcovsky J (2009) A kinase-START gene confers temperature-dependent resistance to wheat stripe rust. Science 323:1357–1360. doi:10.1126/science.1166289
Fukino N, Kunihisa M, Matsumoto S (2004) Characterization of recombinant inbred lines derived from crosses in melon (Cucumis melo L.), ‘PMAR No. 5’ × ‘Harukei No. 3’. Breed Sci 54:141–145
Fukino N, Sakata Y, Kunihisa M, Matsumoto S (2007) Characterisation of novel simple sequence repeat (SSR) markers for melon (Cucumis melo L.) and their use for genotype identification. J Hort Sci Biotechnol 82:330–334
Fukino N, Ohara T, Monforte AJ, Sugiyama M, Sakata Y, Kunihisa M, Matsumoto S (2008a) Identification of QTLs for resistance to powdery mildew and SSR markers diagnostic for powdery mildew resistance genes in melon (Cucumis melo L.). Theor Appl Genet 118:165–175. doi:10.1007/s00122-008-0885-1
Fukino N, Yoshioka Y, Kubo N, Hirai M, Sugiyama M, Sakata Y, Matsumoto S (2008b) Development of 101 novel SSR markers and construction of an SSR-based genetic linkage map of cucumber (Cucumis sativus L.). Breed Sci 58:475–483
Fukino N, Ohara T, Sugiyama M, Kubo N, Hirai M, Sakata Y, Matsumoto S (2012) Mapping of a gene that confers short lateral branching (slb) in melon (Cucumis melo L.). Euphytica 187:133–143. doi:10.1007/s10681-012-0667-3
Fukuoka S, Okuno K (2001) QTL analysis and mapping of pi21, a recessive gene for field resistance to rice blast in Japanese upland rice. Theor Appl Genet 103:185–190
Gonzalo MJ, Oliver M, Garcia-Mas J, Monfort A, Dolcet-Sanjuan R, Katzir N, Arús P, Monforte AJ (2005) Simple-sequence repeat markers used in merging linkage maps of melon (Cucumis melo L.). Theor Appl Genet 110:802–811. doi:10.1007/s00122-004-1814-6
Horejsi T, Staub JE, Thomas C (2000) Linkage of random amplified polymorphic DNA markers to downy mildew resistance in cucumber (Cucumis sativus L.). Euphytica 115:105–113
Huang S, Li R, Zhang Z et al (2009) The genome of the cucumber, Cucumis sativus L. Nat Genet 41:1275–1281. doi:10.1038/ng.475
Kang H, Weng Y, Yang Y, Zhang Z, Zhang S, Mao Z, Cheng G, Gu X, Huang S, Xie B (2011) Fine genetic mapping localizes cucumber scab resistance gene Ccu into an R gene cluster. Theor Appl Genet 122:795–803. doi:10.1007/s00122-010-1487-2
Kong Q, Xiang C, Yu Z (2006) Development of EST-SSRs in Cucumis sativus from sequence database. Mol Ecol Notes 6:1234–1236. doi:10.1111/j.1471-8286.2006.01500.x
Kong Q, Xiang C, Yu Z, Zhang C, Liu F, Peng C, Peng X (2007) Mining and charactering microsatellites in Cucumis melo expressed sequence tags from sequence database. Mol Ecol Notes 7:281–283. doi:10.1111/j.1471-8286.2006.01580.x
Kosambi D (1943) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Kukita Y, Hayashi K (2002) Multicolor post-PCR labeling of DNA fragments with fluorescent ddNTPs. Biotechniques 33:502–506. doi:91200217
Lander E, Green P, Abrahamson J, Barlow A, Daly M, Lincoln S, Newberg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1:174–181
Lehmann EL (1975) Nonparametrics. McGraw-Hill, New York
Liu L, Cai R, Yuan X, He H, Pan J (2008a) QTL molecular marker location of powdery mildew resistance in cucumber (Cucumis sativus L.). Sci China, Ser C Life Sci 51:1003–1008. doi:10.1007/s11427-008-0110-0
Liu L, Yuan X, Cai R, Pan J, He H, Yuan L, Guan Y, Zhu L (2008b) Quantitative trait loci for resistance to powdery mildew in cucumber under seedling spray inoculation and leaf disc infection. J Phytopathol 156:691–697. doi:10.1111/j.1439-0434.2008.01427.x
Miao H, Zhang S, Wang X, Zhang Z, Li M, Mu S, Cheng Z, Zhang R, Huang S, Xie B, Fang Z, Zhang Z, Weng Y, Gu X (2011) A linkage map of cultivated cucumber (Cucumis sativus L.) with 248 microsatellite marker loci and seven genes for horticulturally important traits. Euphytica 182:167–176. doi:10.1007/s10681-011-0410-5
Morishita M, Sugiyama K, Saito T, Sakata Y (2003) Powdery mildew resistance in cucumber. Jpn Agr Res Q 37:7–14
Park Y, Katzir N, Brotman Y, King J, Bertrand F, Havey M (2004) Comparative mapping of ZYMV resistances in cucumber (Cucumis sativus L.) and melon (Cucumis melo L.). Theor Appl Genet 109:707–712. doi:10.1007/s00122-004-1684-y
Ren Y, Zhang Z, Staub J, Cheng Z, Li X, Lu J, Miao H, Kang H, Xie B, Gu X (2009) An integrated genetic and cytogenetic map of the cucumber genome. PLoS ONE 4:1–8. doi:10.1371/journal.pone.0005795
Sakata Y, Kubo N, Morishita M, Kitadani E, Sugiyama M, Hirai M (2006) QTL analysis of powdery mildew resistance in cucumber (Cucumis sativus L.). Theor Appl Genet 112:243–250. doi:10.1007/s00122-005-0121-1
Sakata Y, Morishita M, Kitadani E, Sugiyama M, Ohara T, Sugiyama K, Kojima A (2008) Development of a powdery mildew resistant cucumber (Cucumis sativus L.), ‘Kyuri Chukanbohon Nou 5 Go’. Hort Res (Japan) 7:173–179
van Ooijen JW (2004) MapQTL® 5, Software for the mapping of quantitative trait loci in experimental populations. Kyazma B. V., Wageningen, The Netherlands
Wang S, Basten CJ, Zeng ZB (2007) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC. http://statgen.ncsu.edu/qtlcart/WQTLCart.htm
Watcharawongpaiboon N, Chunwongse J (2008) Development and characterization of microsatellite markers from an enriched genomic library of cucumber (Cucumis sativus). Plant Breed 127:74–81. doi:10.1111/j.1439-0523.2007.01425.x
Weng Y, Staub J, Johnson S, Huang S (2010) An extended intervarietal microsatellite linkage map of cucumber, Cucumis sativus L. Hort Sci 45:882–886
Yamanaka N, Watanabe S, Toda K, Hayashi M, Fuchigami H, Takahashi R, Harada K (2005) Fine mapping of the FT1 locus for soybean flowering time using a residual heterozygous line derived from a recombinant inbred line. Theor Appl Genet 110:634–639. doi:10.1007/s00122-004-1886-3
Yuan XJ, Pan JS, Cai R, Guan Y, Liu LZ, Zhang WW, Li Z, He HL, Zhang C, Si LT, Zhu LH (2008) Genetic mapping and QTL analysis of fruit and flower related traits in cucumber (Cucumis sativus L.) using recombinant inbred lines. Euphytica 164:473–491. doi:10.1007/s10681-008-9722-5
Zeng ZB (1993) Theoretical basis for separation of multiple linked gene effects in mapping quantitative trait loci. Proc Natl Acad Sci USA 90:10972–10976
Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1468
Zhang WW, Pan JS, He HL, Zhang C, Li Z, Zhao JL, Yuan XJ, Zhu LH, Huang SW, Cai R (2012) Construction of a high density integrated genetic map for cucumber (Cucumis sativus L.). Theor Appl Genet 124:249–259. doi:10.1007/s00122-011-1701-x
Acknowledgments
We thank Dr. M. Yano for his valuable advice and Dr. S. Huang for kindly providing SSR marker information prior to publication. We are grateful to S. Negoro, T. Yamakawa, K. Takeuchi and S. Toyoda for their technical assistance. This work was supported by NARO Research Project No. 211 ‘Establishment of Integrated Basis for Development and Application of Advanced Tools for DNA Marker-Assisted Selection in Horticultural Crops’.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11032_2013_9867_MOESM1_ESM.pdf
ESM 1 Comparison of marker distribution and linkage group lengths in the maps from 3 studies: hs map (Ren et al. 2009)a, ss map (Miao et al. 2011)b and CS map (this study)c (PDF 49 kb)
11032_2013_9867_MOESM2_ESM.pdf
ESM 2 Comparison of marker order and map distance in the linkage maps constructed by using RILs derived from inter-varietal (center: hs map, Ren et al. 2009) and intra-varietal crosses (left: ss map, Miao et al. 2011; right: CS map, this study). Common markers are connected with lines. Ruler on the left indicates linkage group length (PDF 427 kb)
11032_2013_9867_MOESM3_ESM.pdf
ESM 3 Positions and LOD scores of QTL pm4.1 for powdery mildew resistance in 109 RILs derived from C. sativus CS-PMR1 × ‘Santou’. Horizontal lines represent LOD thresholds at the 0.05 level of significance. LD25 and LD20, leaf disc assays at 25 and 20 °C, respectively; GH-S and GH-A, evaluation of naturally occurring disease in late spring to summer and in autumn, respectively (PDF 353 kb)
Rights and permissions
About this article
Cite this article
Fukino, N., Yoshioka, Y., Sugiyama, M. et al. Identification and validation of powdery mildew (Podosphaera xanthii)-resistant loci in recombinant inbred lines of cucumber (Cucumis sativus L.). Mol Breeding 32, 267–277 (2013). https://doi.org/10.1007/s11032-013-9867-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-013-9867-3