Abstract
Roots are essential for normal growth, development, and reproduction of higher plants. Consequently, improvement of root system architecture and functionality is of fundamental importance in crop improvement. However, the genetic mechanisms controlling root morphology and function are still not well understood, especially in common wheat, which possesses a complex and unsequenced hexaploid genome. Here we report a more detailed genetic analysis of qTaLRO-B1, a major quantitative trait locus (QTL) previously detected to affect root length and related traits in common wheat. A pair of QTL isolines with different qTaLRO-B1 alleles was developed. Line 178B, carrying the longer root allele, was significantly more efficient in taking up phosphate nutrient and biomass accumulation than line 178A, with the shorter root allele. We mapped qTaLRO-B1 to a 0.9-cM interval on common wheat chromosome 2BS with seven sequence-tagged-site (STS) markers developed from the genes conserved between wheat and Brachypodium distachyon. The seven STS markers were collinearly conserved in tetraploid wheat, but they covered a much larger genetic distance (22.8 cM) in the latter species. In conclusion, we have converted qTaLRO-B1 into a major gene that affects common wheat root length in a qualitative manner, and improved understanding of the genetic location of qTaLRO-B1 and the chromosomal segment carrying this important locus. The implications of our data for further study of qTaLRO-B1 are discussed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Wheat is one of the most important food crops in the world. However, the current annual rate of increase in wheat productivity (about 1.1 %) is considerably below that (≥1.7 %) demanded by global wheat consumption (Dixon 2009; Rosegrant and Agcaoili 2010; Reynolds et al. 2012). Furthermore, wheat production in many regions of the world still relies on the use of excessive water and chemical fertilizers, which is not sustainable either ecologically or economically (Rosegrant et al. 2009; Good and Beatty 2011). Consequently, there is an urgent need to raise the yield potential of wheat and its efficiencies of using water and fertilizer resources. To fulfill this task, the morphology and function of the wheat root system must be improved.
In the past, crop varieties have been bred generally by selection of above-ground organs, whereas the below-ground roots were largely neglected (Dunbabin et al. 2003; Waines and Ehdaie 2007; Den Herder et al. 2010). However, more and more studies show that root development and function, as well as the genes involved, can differ substantially among different genotypes, which can be harnessed for raising water and nutrient use efficiencies and yield potential of crop plants (Coudert et al. 2010; Den Herder et al. 2010). For example, Wasson et al. (2012) recently proposed that, by using appropriate germplasm and combining with suitable laboratory and field screens, it is possible to develop wheat varieties with a deeper and more active root system for achieving higher yield levels in water limited environments. White et al. (2013) suggested a series of morphological and physiological traits for enhancing root uptake of various types of mineral nutrients through breeding. Despite these suggestions, the selection for root traits is difficult at present. In general, root traits are controlled by polygenes with quantitative effects, highly prone to environmental influences, and difficult to observe and quantify under field conditions (Tuberosa et al. 2002a, b; Malamy 2005; Wasson et al. 2012; Hamada et al. 2012). To overcome these problems, large efforts have been devoted to studying the genes and quantitative trait loci (QTLs) controlling root traits in the hope that the resultant resources can facilitate the improvement of root morphology and function through molecular marker-assisted selection (Coudert et al. 2010; Den Herder et al. 2010; Jung and McCouch 2013).
In common wheat, molecular mapping of root trait QTLs have been reported by a number of studies (Sanguineti et al. 2007; Landjeva et al. 2008; Sharma et al. 2011; Hamada et al. 2012; Ren et al. 2012; Bai et al. 2013; Christopher et al. 2013; Liu et al. 2013). Collectively, these studies have demonstrated the presence of multiple QTLs for several major root traits, such as seminal root angle (SRA), number (SRN), and elongation rate (SRER), primary root length (PRL), maximum root length (MRL), lateral root length (LRL) and number (LRN), total root length (TRL), and root surface area (RSA). Furthermore, it has been generally realized that the development and morphological characteristics of seminal roots at seedling stage have important influences on the function of the root system in mature wheat plants. For example, the number of seminal roots has been found to correlate strongly with seed size and grain yield (MacKey 1979; Liu et al. 2013) and to contribute significantly to water uptake (Manschadi et al. 2008) in common wheat. Thus, identifying and analyzing QTLs controlling seminal root traits will likely provide important information and resources for improving root function and its contribution to wheat yield increase. In accordance with this realization, we have previously identified a number of QTLs for seminal root traits using recombinant inbred lines (RILs) derived from two elite common wheat varieties (Xiaoyan 54 and Jing 411; Ren et al. 2012). One QTL uncovered by us, qTaLRO-B1, resided on the short arm of chromosome 2B and explained a large proportion of the phenotypic variation of MRL (68 %) and PRL (59 %). Moreover, through marker allele-assisted genotyping in parental lines and F2 populations, this QTL was detected, and found active, in two other common wheat varieties. Together, these results suggest that qTaLRO-B1 is a major genetic determinant of root length, and may be useful for understanding and improving the genetic basis of root size and function in common wheat. Therefore, it is worthwhile conducting further genetic analysis of qTaLRO-B1.
From the information above, the main objective of this work was to map the chromosomal location of qTaLRO-B1 in more detail and to shed new light on the chromosomal region hosting qTaLRO-B1. Towards this end, a pair of QTL isolines (178A and 178B) segregating for qTaLRO-B1 alleles was developed, and new sequence-tagged-site (STS) markers were developed from the genes conserved between Brachypodium distachyon and wheat. The common wheat chromosomal interval hosting qTaLRO-B1 was compared to its orthologous regions in tetraploid wheat and B. distachyon. The new understanding obtained for qTaLRO-B1 and its implications for further research of qTaLRO-B1 are discussed.
Materials and methods
Development of QTL isolines and comparison of their growth performance
178A and 178B were developed using the heterogeneous inbred family analysis method (Tuinstra et al. 1997). Briefly, immediately after identifying qTaLRO-B1 in the RIL population of Xiaoyan 54 and Jing 411 (Ren et al. 2012), the DNA markers flanking this QTL (i.e., Xgwm210 and Xbarc1138.2) were used to screen the F5 progenies that were accumulated while developing the RILs. Several F5 plants were found to be heterozygous at the qTaLRO-B1 locus. After selfing one such F5 plant, two F6 lines, homozygous for the qTaLRO-B1 alleles from Jing 411 or Xiaoyan 54, were identified through marker-assisted selection (MAS). After three rounds of selfing and MAS, the two lines were designated 178A and 178B, respectively, and used in the experiments described in this study.
Pot culture trail was carried out to compare the root growth and function of 178A and 178B. The soil used for the trail was calcareous, and its available phosphorus (Olsen P) content was 1.6 mg kg−1. The Olsen P content of the soil was adjusted to two different levels, 26 mg kg−1 (high P treatment) and 8 mg kg−1 (low P treatment), by adding the required amounts of KH2PO4. The soil was also supplied with 75 mg kg−1 nitrogen (as urea), 1.0 mg kg−1 zinc (as ZnSO4), 1.0 mg kg−1 boron (as H3BO3), and appropriate quantities of KCl to balance the potassium level. Two uniformly germinated seedlings were planted in each pot, with four replicates arranged per P level per genotype. At 4 weeks after sowing, shoot dry weight, root dry weight, and shoot P content were investigated for every pot. The above- and below-ground parts of the plants in each pot were collected and dried at 80 °C for determining shoot and root dry weight. Shoot P content was measured using the molybdate-blue colorimetric method (Murphy and Riley 1962). Two independent pot culture trails were conducted, with identical results obtained.
Preparation of genetic populations and evaluation of root phenotypes
For investigating the genetic control of the long root phenotype in 178B as well as the mapping of qTaLRO-B1, 57 F1 seeds were created by crossing 178A with 178B, and a segregating F2 population was developed by selfing 20 F1 plants. To aid comparative genomic analysis of qTaLRO-B1, a tetraploid wheat F2 population was constructed using the durum wheat variety Langdon and a wild emmer wheat accession (WEW270) as parents. The seedling root length of Langdon was substantially longer than that of WEW270.
Previous studies have reported that the root trait QTLs detected in hydroponic culture coincided with those for nutrient uptake and grain yield in common wheat (An et al. 2006; Su et al. 2009; Liu et al. 2013). Thus, in the present study, hydroponic culture was used to investigate root morphological parameters of wheat seedlings in the growth chamber (set at 18 °C, with 60 % relative humidity and 16-h light/8-h dark photoperiod). Seeds were germinated in water for 18 h at 20 °C. The germinated seeds were placed onto a nylon mesh to grow for 6 days in water. After removing residual endosperm materials, the seedlings (supported by the nylon mesh) were transferred to plastic boxes (35 × 25 × 10 cm, each containing 5 L of nutrient solution) to grow for another 12 days. The nutrient solution was refreshed every 3 days. The oxygen content in the nutrient solution was enriched by adding 125 μl of 30 % H2O2. At the end of hydroponic culture, MRL was measured for 178A, 178B, F1, and F2 progenies.
DNA isolation and PCR conditions
Genomic DNA was extracted from seedling leaves using the cetyltrimethylammonium bromide method (Allen et al. 2006). PCR was conducted in a total volume of 20 μl containing 10 mM Tris–HCl, pH 7.5, 50 mM KCl, 1.5 mM MgCl2, 0.2 mM of each of the four dNTPs, 50 ng of each of the two primers, 50–100 ng genomic DNA template, and 0.5 U Taq DNA polymerase. Amplification was performed at 94 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 50–60 °C (adjusted for specific primer set) for 30 s, and 72 °C for 30 s, with a final extension at 72 °C for 10 min. PCR products were separated on 6 % denaturing polyacrylamide gels with a 19:1 ratio of acrylamide:bisacrylamide. Gels were silver-stained and photographed.
Development of new STS markers and linkage analysis
Previous work indicated that qTaLRO-B1 was flanked by the microsatellite markers Xgwm210 and Xbarc1138.2 (Ren et al. 2012). Since Xgwm210 had been located to the distal bin of chromosome 2BS (2BS4-0.84-1.00, http://wheat.pw.usda.gov/GG2/index.shtml), and the counterpart of 2BS4-0.84-1.00 in B. distachyon was on the short arm of chromosome 5 (Bd5S, Liu et al. 2012), the genes annotated for a region of Bd5S (from 0.5 to 2.5 Mb) were used to search wheat expressed sequence tag (EST) databases (http://www.cerealsdb.uk.net/CerealsDB/Documents/DOC_database.php, http://blast.ncbi.nlm.nih.gov/Blast.cgi). The positive hits were then employed for designing gene-specific conserved primers (http://probes.pw.usda.gov/cgi-bin/ConservedPrimers/ConservedPrimers.cgi, You et al. 2009). The designed primer sets were each tested by PCR with the genomic DNA samples of 178A or 178B as template. For the primer sets that yielded PCR polymorphic fragments between 178A and 178B, they were further analyzed by PCR with the genomic DNA samples of Chinese Spring and the nulli-tetrasomic lines of wheat group 2 chromosomes (i.e., N2AT2B, N2BT2D, and N2DT2A; Sears 1954) as template. In this way, the primer sets capable of amplifying polymorphic PCR fragments originating from chromosome 2B were identified, and STS markers specific for chromosome 2B were developed. The newly developed STS markers were all labeled by the superscript “TaLRO-B1” to indicate that they were located in the chromosomal region hosting qTaLRO-B1 (Table 1). The alleles of the STS markers were differentiated using 6 % denaturing PAGE optimized as described previously (Creste et al. 2001). In this gel system, the alleles could be distinguished by either length polymorphism or single strand conformation polymorphism (SSCP).
To accomplish linkage analysis of qTaLRO-B1, 5087 F2 individuals were subject to root phenotyping by hydroponic culture and DNA genotyping by PCR as described above. Apart from the newly developed STS markers, three simple sequence repeat (SSR) markers Xgwm210, Xbarc1138.2, and Xgwm429, previously reported in the vicinity of qTaLRO-B1 (Ren et al. 2012), were also used during genotyping. The phenotype and genotype data obtained were deployed to construct a linkage map of qTaLRO-B1 using the software Joinmap 4.0 (Van Ooijen and Voorrips 2006). Using the same software, the linkage relationships of the newly developed STS markers in tetraploid wheat were investigated with the genotyping data collected from 127 F2 individuals descended from the cross Langdon × WEW270 (see above).
Statistical analysis
All data were calculated using Excel 2010, and the results were presented as mean ± standard error. Statistical comparison of the data was performed with the software IBM SPSS Statistics 19. Chi squared tests for goodness-of-fit were used to compare the observed and theoretically expected segregation ratios in the F2, population as described previously (Chernoff and Lehmann 1954).
Results
Genetic analysis of root system difference between 178A and 178B
When hydroponically cultured seedlings of 178A, 178B, and their F1 hybrid were inspected, the root system of 178B was substantially longer than that of 178A, with all F1 samples exhibiting a longer root phenotype (Fig. 1a). Quantitative comparison of MRL confirmed the above observation (Fig. 1b). In contrast, F2 seedlings exhibited clear segregation of longer and shorter root phenotypes (Fig. 1a). The segregation ratio between the two root phenotypes was examined in a subsample of 1,933 F2 seedlings derived from 178A × 178B. The numbers of F2 individuals with longer or shorter roots were 1,458 and 475, respectively, which fitted a 3:1 segregation ratio according to the Chi squared test (χ2 = 0.021, χ 20.05, 1 = 3.84). This segregation ratio, plus the root phenotype of the F1 seedlings (Fig. 1a), indicated that the longer root phenotype displayed by 178B was dominant over the shorter root phenotype exhibited by 178A.
Comparison of the seedling roots of 178A, 178B, and the F1 and F2 individuals derived from 178A × 178B. a Morphological differences between the root systems of typical 178A, 178B, and F1 and F2 plants. The seedlings shown were photographed under the same magnification. b Quantitative differences in maximum root length (MRL) among 178A, 178B, and F1 plants. The data set displayed is representative of three separate measurements. Asterisk indicates statistically significant difference (P < 0.01) from the mean MRL values of 178B and F1 plants
Comparison of growth performance between 178A and 178B
Two independent pot cultivation experiments were carried out to compare the growth of 178A and 178B under the conditions of high and low levels of P supply. In both experiments, considerable differences were found between 178A and 178B in terms of shoot and root dry weight accumulation and shoot P content. Generally, the values determined for 178B were significantly higher than those for 178A (Fig. 2).
Comparison of shoot dry weight, root dry weight, and shoot phosphorus (P) content between 178A and 178B under high P (HP) or low P (LP) growth conditions. The data set shown is representative of two separate pot culture experiments. Asterisk indicates statistically significant difference (P < 0.05) between the measurements of 178A and 178B
Further genetic mapping of qTaLRO-B1
A prerequisite for further mapping of qTaLRO-B1 was the finding of DNA markers that were polymorphic between 178A and 178B and yet located in the chromosomal bin 2BS4-0.84-1.00 where qTaLRO-B1 resided (see “Materials and methods”). We tested a wide range of SSR and EST markers previously found in 2BS4-0.84-1.00, but none of them showed polymorphism between 178A and 178B. Consequently, efforts were made to develop new STS markers from the genes syntenically conserved between the distal part of wheat 2BS chromosome and its orthologous region on the short arm of chromosome 5 of B. distachyon (Bd5S, Liu et al. 2012). Although more than 120 genes from Bd5S were used to search conserved wheat ESTs, and nearly 100 pairs of conserved primers were designed, only seven primer sets amplified polymorphic PCR products that were confirmed to be located on the distal region of 2BS by analysis with Chinese Spring, N2AT2B, N2BT2D, and N2DT2A. The seven primer sets represented STS markers newly developed by this work, and were designated STS1 TaLRO-B1 to STS7 TaLRO-B1 (Table 1). Except for STS3 TaLRO-B1, which was a dominant marker, the remaining six were all co-dominant for 178A and 178B (Table 1). The alleles of STS1 TaLRO-B1, STS3 TaLRO-B1, and STS4 TaLRO-B1 were differentiated by length polymorphism, whereas those of the remaining four markers were distinguished by SSCP (Table 1; Supplementary Fig. 1).
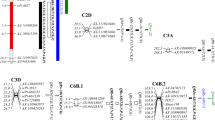
With the genotyping and MRL data from 5087 F2 individuals, a linkage map consisting of 10 DNA markers and qTaLRO-B1 was constructed (Fig. 3). The total genetic distance covered by this regional map was 58.7 cM. STS5 TaLRO-B1 and STS6 TaLRO-B1 were the closest flanking markers of qTaLRO-B1, and their genetic distances to qTaLRO-B1 were 0.1 and 0.3 cM, respectively (Fig. 3). The distances of qTaLRO-B1 to Xgwm210 and Xbarc1138.2 were 0.4 and 0.2 cM, respectively (Fig. 3). Finally, it was observed that STS2 TaLRO-B1, STS3 TaLRO-B1, STS4 TaLRO-B1, and Xbarc1138.2 were genetically linked together (Fig. 3), despite the analysis of more than 5000 F2 plants.
Genetic linkage map of the region carrying qTaLRO-B1 on the short arm of chromosome 2B (2BS) of common wheat, constructed with seven STS (STS1 TaLRO-B1–STS7 TaLRO-B1) and three microsatellite (Xgwm210, Xbarc1138, and Xgwm429) markers. The genetic distance value (cM) is shown in the left panel. Arrow indicates the position of qTaLRO-B1
Comparison of qTaLRO-B1 region and its orthologous counterparts in tetraploid wheat and B. distachyon
For this set of analyses, we concentrated on the newly developed STS markers as they were more likely to be collinearly conserved among the three species. In tetraploid wheat, the loci corresponding to STS1 TaLRO-B1 and STS7 TaLRO-B1 were all conserved. However, STS4 TaLRO-B1 could not be mapped because it was not polymorphic between the parents of the tetraploid wheat F2 population developed in this work. For the six markers that were mapped in tetraploid wheat, their genetic positions were collinear to those found in common wheat (Fig. 4). Interestingly, the genetic distance value from STS1 TaLRO-B1 to STS7 TaLRO-B1 was 22.8 cM, which was approximately 25.3 times larger than that covered by the two markers in common wheat (Fig. 4). Moreover, STS2 TaLRO-B1 and STS3 TaLRO-B1 were separated in tetraploid wheat, in contrast to their tight linkage in common wheat (Fig. 4). Although MRL also segregated among the F2 progenies in the tetraploid population, the gene involved was not genetically linked to any of the seven STS markers (data not shown).
Comparative genomic analysis of the 2BS chromosomal region hosting qTaLRO-B1 and its orthologous counterparts located on the 2BS chromosome of tetraploid wheat or the short arm of chromosome 5 of B. distachyon (Bd5S). Orthologous loci are connected by dashed lines. STS4 TaLRO-B1 could not be mapped in the tetraploid wheat population developed by this work owing to lack of polymorphism between the parental lines. The total genetic distance value from STS1 TaLRO-B1 to STS7 TaLRO-B1, as shown in the parentheses, was substantially larger in tetraploid wheat (22.8 cM) than in common wheat (0.9 cM). The total physical distance covered by the seven B. distachyon gene loci, as indicated in the square brackets, was approximately 1.62 Mb
In B. distachyon, the loci corresponding to STS1 TaLRO-B1 and STS7 TaLRO-B1, as represented by seven annotated genes, were also conserved, but the chromosomal segment corresponding to that covered from STS2 TaLRO-B1 and STS6 TaLRO-B1 in wheat was inverted (Fig. 4). The physical distance spanned by the seven B. distachyon gene loci was approximately 1.62 Mb, whereas that between Bd5g01280 and Bd5g01010, which were orthologous to STS5 TaLRO-B1 and STS6 TaLRO-B1, respectively, was about 0.25 Mb (Fig. 4).
Discussion
Conversion of qTaLRO-B1 into a major gene controlling root growth and function in common wheat
Prior to this work, many QTLs controlling root traits have been identified in common wheat (see “Introduction”) and other crop species (e.g., Zhang et al. 2001; Tuberosa et al. 2002a, b; Fita et al. 2008; Liang et al. 2010; Obara et al. 2010; Ron et al. 2013; Topp et al. 2013), indicating that genetic and functional dissection of QTLs is becoming an essential avenue for understanding the genes involved in root growth and function of crop plants. However, for more advanced characterization of an important QTL, it is often necessary to convert it into a major gene with qualitative effect on its target trait (Shen et al. 2001; Tuberosa and Salvi 2007). Therefore, we developed the QTL isolines 178A and 178B, and investigated whether qTaLRO-B1 might behave as a major gene to control root length in a Mendelian fashion in the genetic population derived from 178A × 178B. From the data shown in Figs. 1 and 2, it is clear that the phenotypic difference between the root systems of 178A and 178B seedlings was caused by allelic variation of a single major gene, and that the root system of 178B was not only longer (in length) and larger (in biomass), but also more active (in inorganic P uptake), than that of 178A. Under our hydroponic culture conditions, the total root surface area (RSA) of 178B (69.7 ± 7.1 cm2) was also significantly higher than that of 178A (36.7 ± 2.7 cm2). The longer length, larger biomass, and higher RSA of 178B’s root system agree well with the enhanced P accumulation found in this genotype relative to its isoline 178A (Fig. 2). Because 178A and 178B were isolines derived from a single F5 plant and differed mainly at the qTaLRO-B1 locus, we suggest that the difference between the root systems of 178A and 178B is controlled by allelic variation of qTaLRO-B1, and that the long root (LR) allele in 178B (originally from Xiaoyan 54) is dominant over the short root (SR) allele in 178A (originally from Jing 411).
The contrasting effects conferred by the LR or SR alleles of qTaLRO-B1 are consistent with previous observations on the possession of larger root biomass and higher phosphorus uptake capacity by Xiaoyan 54 but not Jing 411 (Yan et al. 2010). Since Xiaoyan 54 has also been found to be efficient in acquiring nitrogen (N) from soil (Zhang et al. 2005), we are now in the process of investigating whether 178B has a higher N uptake rate than 178A. As several major root trait QTLs have been found to affect grain production (Landi et al. 2007, 2010; Ehdaie et al. 2010; Suji et al. 2012), it will also be important in further research to test whether 178A and 178B differ in yield level.
Improved understanding of qTaLRO-B1 and the chromosomal region carrying qTaLRO-B1
Although our earlier work detected qTaLRO-B1 in common wheat, knowledge of the genetic location of this locus and the chromosomal segment carrying it was limited (Ren et al. 2012). Based on the linkage mapping and comparative genomic analysis data collected in this work, we have improved understanding of qTaLRO-B1 in three respects. First, with the development and mapping of seven STS markers, the genetic location of qTaLRO-B1 in 2BS is now better defined than before. Second, judging from the collinearity between the seven STS markers and their orthologs in tetraploid wheat, there is a high level of synteny between the qTaLRO-B1 region in common wheat and its orthologous counterpart in tetraploid wheat, although no MRL controlling gene was detected in the latter by this work. The loci corresponding to the seven STS markers are also conserved in B. distachyon. However, the order of these loci in B. distachyon shows some differences from that in common wheat because of a local inversion occurring in the relevant chromosomal region in the former species. Finally, the recombination frequency of the chromosomal region carrying qTaLRO-B1 is substantially lower than that of its syntenic counterpart in tetraploid wheat. This is supported by (1) a large difference between the genetic distances covered by the same set of markers in common wheat and tetraploid wheat, and (2) a clear separation of STS2 TaLRO-B1 from STS3 TaLRO-B1 in tetraploid wheat but not common wheat.
The low recombination frequency in the qTaLRO-B1 region, as compared to that of its syntenic segment in tetraploid wheat, is very likely caused by the suppression of meiotic recombination. This phenomenon has been found for a number of chromosomal regions harboring important genes in plants. For example, recombination frequency is low in the region carrying Pi-ta, a gene conferring strong resistance to Magnaporthe oryzae in rice (Jia 2009; Jia et al. 2012). In wheat, suppressed recombination has been found in the genomic region hosting Lr20-Pm1 conferring resistance to leaf rust and powdery mildew diseases (Neu et al. 2002), and the chromosomal region carrying Ppd-B1 regulating photoperiod response (Mohler et al. 2004). Although the molecular basis of recombination suppression in plants is still not completely resolved (Jia et al. 2012), two possible mechanisms are frequently discussed. The first is that the presence of complex chromosomal structure or rearrangements tends to lead to recombination suppression (Gill et al. 1996a, b; Neu et al. 2002; Kim et al. 2005). The second mechanism is that strong natural or artificial selections may result in certain linkage blocks that have greatly suppressed recombination (Yonemaru et al. 2012). Since qTaLRO-B1 confers positive benefits on the growth of both root and above-ground organs, and is detected in different elite common wheat varieties (Ren et al. 2012; this work), it is possible that the qTaLRO-B1 region has experienced enhanced artificial selection, and thus exhibits substantially lowered recombination frequency. As an initial test for this possibility, we investigated the allelic variation of STS2 TaLRO-B1, STS3 TaLRO-B1, and STS4 TaLRO-B1 in 94 elite common wheat varieties released mainly from the 1980s to 2000s (Zhang et al. 2013). The number of varieties having 178B-type alleles in the three loci was 60, whereas that with 178A-type alleles across these loci was only nine, with the remaining varieties carrying alternative allele(s) in one or more of the three loci (data not shown). This provides a preliminary support for the existence of strong artificial selection for the qTaLRO-B1 region. Molecular cloning of qTaLRO-B1 will enable a more detailed examination of this possibility in the future.
Implications for further research of qTaLRO-B1
From the discussions above, it is apparent that substantial efforts are still needed to fine-map, and to clone, qTaLRO-B1. Due to recombination suppression, the populations derived from 178A × 178B are not suitable for fine mapping of qTaLRO-B1. Consequently, more genetic populations involving 178B are being prepared and evaluated. Concomitantly, additional STS markers are being developed by comparing B. distachyon genes located between Bd5g01280 and Bd5g01010 (which corresponded to STS5 TaLRO-B1 and STS6 TaLRO-B1, respectively; Fig. 4) with wheat ESTs. The application of these new resources should enable further improvement in understanding the qTaLRO-B1 region, and may help to identify more DNA markers and donor genotypes suitable for transferring qTaLRO-B1 in MAS breeding aimed at improving common wheat root size and function.
References
Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF (2006) A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc 1:2320–2325
An DG, Su JY, Liu QY, Li B, Jing RL, Li JY, Li ZS (2006) Mapping QTLs for nitrogen uptake in relation to the early growth of wheat (Triticum aestivum L.). Plant Soil 284:73–84
Bai C, Liang Y, Hawkesford MJ (2013) Identification of QTLs associated with seedling root traits and their correlation with plant height in wheat. J Exp Bot 64:1745–1753
Chernoff H, Lehmann EL (1954) The use of maximum likelihood estimates in χ2 tests for goodness-of-fit. Ann Math Stat 25:579–586
Christopher J, Christopher M, Jennings R, Jones S, Fletcher S, Borrell A, Manschadi AM, Jordan D, Mace E, Hammer G (2013) QTL for root angle and number in a population developed from bread wheats (Triticum aestivum) with contrasting adaptation to water-limited environments. Theor Appl Genet 126:1563–1574
Coudert Y, Périn C, Courtois B, Khong NG, Gantet P (2010) Genetic control of root development in rice, the model cereal. Trends Plant Sci 15:219–226
Creste S, Neto T, Figueira A (2001) Detection of single sequence repeat polymorphisms in denaturing polyacrylamide sequencing gels by silver staining. Plant Mol Biol Rep 19:299–306
Den Herder G, Van Isterdael G, Beeckman T, De Smet I (2010) The roots of a new green revolution. Trends Plant Sci 15:600–607
Dixon JM (2009) Wheat facts and futures 2009. CIMMYT, Mexico
Dunbabin V, Diggle A, Rengel Z (2003) Is there an optimal root architecture for nitrate capture in leaching environments? Plant Cell Environ 26:835–844
Ehdaie B, Merhaut DJ, Ahmadian S, Hoops AC, Khuong T, Layne AP, Waines JG (2010) Root system size influences water-nutrient uptake and nitrate leaching potential in wheat. J Agron Crop Sci 196:455–466
Fita A, Picó B, Monforte AJ, Nuez F (2008) Genetics of root system architecture using near-isogenic lines of melon. J Am Soc Hort Sci 133:448–458
Gill KS, Gill BS, Endo TR, Boyko EV (1996a) Identification and high-density mapping of gene rich regions in chromosome group 5 of wheat. Genetics 143:1001–1012
Gill KS, Gill BS, Endo TR, Taylor T (1996b) Identification and high-density mapping of gene rich regions in chromosome group 1 of wheat. Genetics 144:1883–1891
Good A, Beatty P (2011) Biotechnological approaches to improving nitrogen use efficiency in plants: alanine aminotransferase as a case study. In: Hawkesford MJ, Barraclough P (eds) The molecular and physiological basis of nutrient use efficiency in crops. Wiley-Blackwell, Oxford. doi:10.1002/9780470960707.ch9
Hamada A, Nitta M, Nasuda S, Kato K, Fujita M, Matsunaka H, Okumoto Y (2012) Novel QTLs for growth angle of seminal roots in wheat (Triticum aestivum L.). Plant Soil 354:395–405
Jia Y (2009) Artificial introgression of a large fragment around the Pi-ta rice blast resistance gene in backcross progenies and several elite rice cultivars. Heredity 103:333–339
Jia Y, Jia MH, Wang Z, Liu G (2012) Indica and Japonica crosses resulting in linkage block and recombination suppression on rice chromosome 12. PLoS ONE 7:e43066
Jung JK, McCouch S (2013) Getting to the roots of it: genetic and hormonal control of root architecture. Front Plant Sci 4:186
Kim JS, Islam-Faridi MN, Klein PE, Stelly DM, Price HJ, Klein RR, Mullet JE (2005) Comprehensive molecular cytogenetic analysis of sorghum genome architecture: distribution of euchromatin, heterochromatin, genes and recombination in comparison to rice. Genetics 171:1963–1976
Landi P, Sanguineti MC, Liu C, Li Y, Wang TY, Giuliani S, Bellotti M, Salvi S, Tuberosa R (2007) Root-ABA1 QTL affects root lodging, grain yield and other agronomic traits in maize grown under well-watered and water-stressed conditions. J Exp Bot 58:319–326
Landi P, Giuliani S, Salvi S, Ferri M, Tuberosa R, Sanguineti MC (2010) Characterization of root-yield-1.06, a major constitutive QTL for root and agronomic traits in maize across water regimes. J Exp Bot 61:3553–3562
Landjeva S, Neumann K, Lohwasser U, Börner A (2008) Molecular mapping of genomic regions associated with wheat seedling growth under osmotic stress. Biol Plant 52:259–266
Liang Q, Cheng X, Mei M, Yan X, Liao H (2010) QTL analysis of root traits as related to phosphorus efficiency in soybean. Ann Bot 106:223–234
Liu Z, Zhu J, Cui Y, Liang Y, Wu H, Song W, Liu Q, Yang T, Sun Q, Liu Z (2012) Identification and comparative mapping of a powdery mildew resistance gene derived from wild emmer (Triticum turgidum var. dicoccoides) on chromosome 2BS. Theor Appl Genet 124:1041–1049
Liu X, Li R, Chang X, Jing R (2013) Mapping QTLs for seedling root traits in a doubled haploid wheat population under different water regimes. Euphytica 189:51–66
MacKey J (1979) Genetic potentials for improved yield. Acta Agron Hung 28:121–143
Malamy JE (2005) Intrinsic and environmental response pathways that regulate root system architecture. Plant, Cell Environ 28:67–77
Manschadi AM, Hammer GL, Christopher JT, de Voil P (2008) Genotypic variation in seedling root architectural traits and implications for drought adaptation in wheat (Triticum aestivum L.). Plant Soil 303:115–129
Mohler V, Lukman R, Ortiz-Islas S, William M, Worland AJ, van Beem J, Wenzel G (2004) Genetic and physical mapping of photoperiod insensitive gene Ppd-B1 in common wheat. Euphytica 138:33–40
Murphy JR, Riley JP (1962) A modified single solution method for the determination of phosphate in natural waters. Anal Chim Acta 27:31–36
Neu C, Stein N, Keller B (2002) Genetic mapping of the Lr20-Pm1 resistance locus reveals suppressed recombination on chromosome arm 7AL in hexaploid wheat. Genome 45:737–744
Obara M, Tamura W, Ebitani T, Yano M, Sato T, Yamaya T (2010) Fine-mapping of qRL6.1, a major QTL for root length of rice seedlings grown under a wide range of NH4+ concentrations in hydroponic conditions. Theor Appl Genet 121:535–547
Ren Y, He X, Liu D, Li J, Zhao X, Li B, Tong Y, Zhang A, Li Z (2012) Major quantitative trait loci for seminal root morphology of wheat seedlings. Mol Breed 30:139–148
Reynolds M, Foulkes J, Furbank R, Griffiths S, King J, Murchie E, Parry M, Slafer G (2012) Achieving yield gains in wheat. Plant Cell Environ 35:1799–1823
Ron M, Dorrity MW, de Lucas M, Toal T, Hernandez RI, Little SA, Maloof JN, Kliebenstein DJ, Brady SM (2013) Identification of novel loci regulating interspecific variation in root morphology and cellular development in tomato. Plant Physiol 162:755–768
Rosegrant MW, Agcaoili M (2010) Global food demand, supply, and price prospects to 2010. International Food Policy Research Institute, Washington
Rosegrant MW, Ringler C, Zhu T (2009) Water for agriculture: maintaining food security under growing scarcity. Annu Rev Environ Res 34:205–222
Sanguineti MC, Li S, Maccaferri M, Corneti S, Rotondo F, Chiari T, Tuberosa R (2007) Genetic dissection of seminal root architecture in elite durum wheat germplasm. Ann Appl Biol 151:291–305
Sears ER (1954) The aneuploids of common wheat. MO Agr Exp Sta Res Bull 572. University of Missouri, Columbia
Sharma S, Xu S, Ehdaie B, Hoops A, Clsoe TJ, Lukaszewski AJ, Waines JG (2011) Dissection of QTL effects for root traits using a chromosome arm-specific mapping population in bread wheat. Theor Appl Genet 122:759–769
Shen B, Courtois B, McNally KL, Robin S, Li Z (2001) Evaluation of near-isogenic lines of rice introgressed with QTLs for root depth through marker-aided selection. Theor Appl Genet 103:75–83
Su JY, Zheng Q, Li HW, Li B, Jing RL, Tong YP, Li ZS (2009) Detection of QTLs for phosphorus use efficiency in relation to agronomic performance of wheat grown under phosphorus sufficient and limited conditions. Plant Sci 176:824–836
Suji KK, Prince K, Mankhar PS, Kanagaraj P, Poornima R, Amutha K, Kavitha S, Biji KR, Gomez SM, Babu RC (2012) Evaluation of rice (Oryza sativa L.) near iso-genic lines with root QTLs for plant production and root traits in rainfed target populations of environment. Field Crop Res 137:89–96
Topp CN, Iyer-Pascuzzi AS, Anderson JT, Lee CR, Zurek PR, Symonova O, Zheng Y, Bucksch A, Mileyko Y, Galkovskyi T, Moore BT, Harer J, Edelsbrunner H, Mitchell-Olds T, Weitz JS, Benfey PN (2013) 3D phenotyping and quantitative trait locus mapping identify core regions of the rice genome controlling root architecture. Proc Natl Acad Sci USA 110:1695–1704
Tuberosa R, Salvi S (2007) From QTLs to genes controlling root traits in maize. In: Spiertz JHJ, Struik PC, van Laar HH (eds) Scale and complexity in plant systems research: gene–plant–crop relations. Springer, The Netherlands, pp 13–22
Tuberosa R, Salvi S, Sanguineti MC, Landi P, Maccaferri M, Conti S (2002a) Mapping QTLs regulating morpho-physiological traits and yield: case studies, shortcomings and perspectives in drought-stressed maize. Ann Bot 89:941–963
Tuberosa R, Sanguineti MC, Landi P, Michela Giuliani M, Salvi S (2002b) Identification of QTLs for root characteristics in maize grown in hydroponics and analysis of their overlap with QTLs for grain yield in the field at two water regimes. Plant Mol Biol 48:697–712
Tuinstra MR, Ejeta G, Goldsbrough PB (1997) Heterogeneous inbred family (HIF) analysis: a method for developing near-isogenic lines that differ at quantitative trait loci. Theor Appl Genet 95:1005–1011
Van Ooijen JW, Voorrips RE (2006) JoinMap4.0, software for the calculation of genetic linkage maps. Plant Research International, Wageningen
Waines JG, Ehdaie B (2007) Domestication and crop physiology: roots of green-revolution wheat. Ann Bot 100:991–998
Wasson AP, Richards RA, Chatrath R, Misra SC, Prasad SV, Rebetzke GJ, Kirkegaard JA, Christopher J, Watt M (2012) Traits and selection strategies to improve root systems and water uptake in water-limited wheat crops. J Exp Bot 6:3485–3498
White PJ, George TS, Dupuy LX, Karley AJ, Valentine TA, Wiesel L, Wishart J (2013) Root traits for infertile soils. Front Plant Sci 4:193
Yan HL, Liu WK, Li GH, Zhang SX (2010) Comparison of rhizosphere impacts of wheat genotypes differing in phosphorus efficiency. Can J Plant Sci 90:311–317
Yonemaru JI, Yamamoto T, Ebana K, Yamamoto E, Nagasaki H, Shibaya T, Yano M (2012) Genome-wide haplotype changes produced by artificial selection during modern rice breeding in Japan. PLoS ONE 7:e32982
You FM, Huo N, Gu YQ, Lazo GR, Dvorak J, Anderson OD (2009) Conserved Primers 2.0: a high-throughput pipeline for comparative genome referenced intron-flanking PCR primer design and its application in wheat SNP discovery. BMC Bioinform 10:331
Zhang J, Zheng HG, Aarti A, Pantuwan G, Nguyen TT, Tripathy JN, Sarial AK, Robin S, Babu RC, Nguyen BD et al (2001) Locating genomic regions associated with components of drought resistance in rice: comparative mapping within and across species. Theor Appl Genet 103:19–29
Zhang J, Ju XT, Gao Q, Zhang FS (2005) Recovery of labeled nitrate-N in different soil layers by two crops, spinach and wheat. Sci Agric Sin 38:333–340
Zhang K, Wang J, Zhang L, Rong C, Zhao F, Peng T, Li H, Cheng D, Liu X, Qin H, Zhang A, Tong Y, Wang D (2013) Association analysis of genomic loci important for grain weight control in elite common wheat varieties cultivated with variable water and fertiliser supply. PLoS ONE 8:e57853
Acknowledgments
This research was supported by the Ministry of Science and Technology of China (via grants 2011BAD07B02-2 and 2011CB100300). We thank Professors Junhua Peng and Eviatar Nevo for supplying the wild emmer wheat accession WEW270, and Shuangjuan Yang for technical advice on data processing during linkage analysis.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Pei Cao and Yongzhe Ren have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cao, P., Ren, Y., Zhang, K. et al. Further genetic analysis of a major quantitative trait locus controlling root length and related traits in common wheat. Mol Breeding 33, 975–985 (2014). https://doi.org/10.1007/s11032-013-0013-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-013-0013-z