Abstract
Fusarium wilt caused by Fusarium oxysporum f. sp. conglutinans is one of the most important diseases of Brassica crops, resulting in severe reductions in yield and quality. To characterize the inheritance pattern of fusarium resistance, a cross between a susceptible broccoli and a resistant cabbage was subjected to segregation analysis. Results indicated that resistance was controlled by a single dominant allele. This gene was named Foc-Bo1 and mapped to linkage group seven (O7) by both the segregation test and quantitative trait locus (QTL) analysis. The QTL on O7 was detected with a logarithm of odds score (LOD) of 19.5, which was above the threshold value with genome-wide 1% significance level (2.01). A minor QTL was also detected on O4 with a LOD score of 2.06. Inoculation tests indicated that stable expression of fusarium resistance at high temperatures required Foc-Bo1 homozygosity. The association between Foc-Bo1 and the closest simple sequence repeat marker (KBrS003O1N10) was analyzed in three F3 populations. Based on these studies, KBrS003O1N10 represents an effective marker-assisted selection (MAS) tool for breeding fusarium wilt resistance into Brassica oleracea crops. To our knowledge, this is the first paper to map the fusarium-resistance gene in Brassica species and to validate the effectiveness of MAS in improving fusarium resistance in these important plants.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cole crops derived from the Brassica oleracea L. species include cabbage, broccoli, cauliflower, and Brussels sprouts. These important vegetables are cultivated throughout the world, but most suffer from a damaging disease called fusarium wilt (Ramirez-Villupadua et al. 1985; Morrison et al. 1994; Farnham et al. 2001). Fusarium wilt of Brassica crops is caused by the soil-borne pathogen Fusarium oxysporum f. sp. conglutinans (Wollenweb.) (Snyder and Hansen 1940). Fusarium colonization leads to leaf yellowing, wilting, defoliation (in older plants), stunted growth, and plant death. As a result, crops suffer severe losses in both yield and quality (Walker 1930). Fusarium wilt is a warm-weather disease that is typically not a problem in cool conditions (i.e., cool seasons or environments). However, increased levels of cole crop production in temperate climates and during warm seasons have made this disease a serious problem (Bosland et al. 1988; Farnham et al. 2001). Global warming may also contribute to the rise in fusarium wilt incidence (Berrocal-Lobo and Molina 2007). Crop rotation, improved sanitation, and fungicides are ineffective measures against this disease, as these ubiquitous pathogens cannot be eradicated from the soil. Thus, the development of resistant cultivars is the most cost-effective method for controlling fusarium wilt in Brassica crops.
Cabbage cultivars carrying resistance to fusarium wilt were initially developed in the United States at the beginning of the twentieth century (Walker et al. 1927; Anderson 1933). Progenies obtained from crosses between resistant and susceptible cultivars were subsequently analyzed to determine the genetic basis of fusarium wilt resistance (Walker 1930; Walker and Smith 1930; Blank 1937). Two types of cabbage resistance against fusarium wilt have been reported: Type A and Type B (Blank 1937). Type A resistance is controlled by a single dominant gene and is phenotypically stable at temperatures lower than 26–28°C. In contrast, Type B is polygenic and becomes unstable as temperatures rise above 24°C (Walker and Smith 1930; Blank 1937; Walker 1953). Type A represents the more advantageous form of resistance because it is inherited in a monogenic fashion, which facilitates transfer between varieties, and it is phenotypically stable at soil temperatures as high as 28°C (Walker and Smith 1930; Blank 1937; Walker 1953). For these reasons, Type A resistance has been used effectively in cabbage and is often cited as a successful example of long-lasting, monogenic, dominant resistance (Dixon 2007). Identification of the Type A resistance gene is important to improve fusarium wilt resistance. To date, however, no studies concerning the molecular or genetic basis of fusarium wilt resistance have been conducted in B. oleracea.
The objectives of this study, therefore, were (1) to analyze the inheritance pattern of fusarium resistance in an F2 population of plants, both by segregation tests and quantitative trait locus (QTL) analysis, and (2) to develop a marker-assisted selection (MAS) process for fusarium wilt resistance in B. oleracea.
Materials and methods
Plant materials
Microspore culture was used to obtain double-haploid lines of the fusarium wilt-resistant cabbage cultivar Anju (Nippon Norin Seed Co., Japan), and the fusarium wilt-susceptible broccoli cultivar Green Comet (GC) (Takii & Co., Ltd., Japan). The GC line was crossed to the Anju line to generate the F1 population (GC was used as the female). An F1 plant was randomly selected and self-pollinated to generate 94 F2 progenies of single-plant descent. These F2 plants were used by Nagaoka et al. (2010) to construct a genetic linkage map. F3 seeds were obtained from bud self-pollination of each F2 line.
Pathogen isolation and inocula preparation
The Cong: 1-1 strain of F. oxysporum f. sp. conglutinans obtained from cabbage was provided by Dr. Kadota (National Agricultural Research Center for Tohoku Region, Japan), and was used to prepare inocula. This strain was maintained on potato dextrose agar medium (Wako Pure Chemical Industries, Ltd., Japan). Liquid inocula were obtained by inoculating potato sucrose broth medium (200 g/l potato extract and 20 g/l sucrose in distilled water) with the isolate, and shaking at 130 rpm on a rotary shaker for 1 week. After shaking, 50 ml of liquid inocula was poured into a sterilized soil mixture that contained 80 g of soil mix (Yasaibaido No. 1, Hone Agri Co., Ltd., Japan), 20 g of rice bran powder, and 30 ml of a 4% sucrose solution. The inoculated soil medium was incubated at 25°C for 2 weeks and then used for inoculation tests in both the greenhouse and growth chambers. For growth chamber tests, 2 g of perlite (a soil conditioner that improves soil aeration) was added to the inoculated soil medium. As the inoculated soil medium contained a high dose of conidia from F. oxysporum, a healthy soil mix was added to the inoculated soil medium at transplantation. Ratios of either 4:1 or 9:1 of healthy soil mix to inoculated soil medium were used.
Evaluation of fusarium resistance
Artificial inoculation tests were performed in either a greenhouse or a growth chamber. In the greenhouse, inoculation tests were performed from August 16 to September 5, 2008. The mean temperature in the greenhouse during that time was 26.4 ± 2.1°C (mean ± SD), with a mean daily high temperature of 33.2 ± 3.1°C and a mean daily low temperature of 22.2 ± 1.9°C (Electronic Supplementary Material Fig. S1). The disease severity index (DI) of F2 was evaluated from the average of 12 F3 plants (F2:3). Eighty-five F2:3 populations were screened for fusarium wilt resistance. These lines were selected from the 94 F2 populations that were previously used to develop a linkage map (Nagaoka et al. 2010). Twelve seeds from each F3 family (derived from the 85 F2 plants) were sown in a plastic tray. Twelve-day-old seedlings were transplanted into plastic pots (8 cm in diameter) that contained both the healthy soil mix and the inoculated soil medium (mixed 4:1). Soil was kept moist throughout the test. The DI of each inoculated plant was determined 14 days after transplantation. A DI scale of 0–2 was used (0: no symptoms, 1: plant yellowing, 2: plant death). DIs of the 12 F3 seedlings were averaged to calculate the DI for each F2 plant.
In the growth chambers, 12-day-old seedlings were transplanted into plastic pots (8 cm in diameter). The ratio of the healthy soil mix to the inoculated soil medium was adjusted to 9:1. The DI of each inoculated plant was determined 14 days after transplantation, using the same 0–2 scale that was used for the greenhouse experiments. The growth chamber tests were carried out using three F3 lines (see “Marker validation” in “Materials and methods”) and two temperature regimens: (1) 25°C for 7 days, followed by 28°C for 7 days, or (2) 28°C constant temperature for 14 days.
Detection of DNA polymorphism
Each 5-μl polymerase chain reaction (PCR) mixture contained 0.125 U Takara Ex-Taq (Takara Bio Inc., Otsu, Japan), 0.5 μl 10 × Ex-Taq buffer, 2 nmol of each dNTP, 5 pmol of each primer, and ~10 ng genomic DNA. Amplification was performed using a PCR thermal cycler (Takara Bio). The reaction mixture was incubated in the thermal cycler at 94°C for 5 min, followed by 35 cycles of 94°C for 30 s (denaturation), 50°C for 30 s (annealing), and 72°C for 1 min (elongation). PCR products were separated by electrophoresis using polyacrylamide gel (Kikuchi et al. 2003). Gels were stained with Gelstar solution (diluted 1:10,000) (Takara Bio), and visualized under UV light. Segregation of each marker was scored with reference to parental genotypes. To detect the homologues of RESISTANCE TO FUSARIUM OXYSPORUM 1 (RFO1), which confers resistance to fusarium in Arabidopsis thaliana (Diener and Ausubel 2005), the PCR primers (forward: 5′-GTTACAGTAACTGTTACTGC-3′ designed from exon 1; reverse: 5′-GTTAATCTGCGAGAGAACAC-3′ designed from exon 3) were developed based on the RFO1 gene sequence (At1g79670) and used to amplify the homologous fragments in the B. oleracea genome. Polymorphism detection between the Anju and GC genomes was carried out using the same procedure as above-mentioned.
Segregation test and QTL analysis in F2 plants
DIs of individual F2 plants were deduced from F3 progeny tests. Since a gap appeared in the DI distribution of the F2 population, plants with a DI below 1.4 or above 1.6 were classified as disease-resistant or disease-susceptible, respectively (Fig. 1). The ratio of resistant to susceptible plants in the F2 population was then calculated and analyzed by the χ2 test.
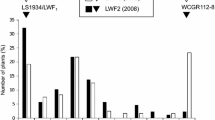
Frequency distribution of the disease severity index (DI) for F2 progeny. Arrows indicate approximate values obtained for parental (Anju and GC) and F1 plants. The DI for each F2 plant was deduced by averaging the DIs of 12 F3 plants. F2 plants homozygous for the Anju KBrS003O10N1 locus, homozygous for the GC KBrS003O10N1 locus, or heterozygous at the KBrS003O10N1 locus are indicated by open, black, or gray bars, respectively
A linkage map was constructed from the 94 F2 population previously described by Nagaoka et al. (2010). The linkage map generated by Nagaoka et al. (2010) was modified in this study by including additional markers in the analysis. Linkage analysis was performed using the Antmap 1.2 program (Iwata and Ninomiya 2006). To identify QTL that contribute to fusarium wilt resistance, an interval mapping method developed by Hayashi and Awata (2006) for the analysis of categorical traits was used, in which the genomic regions (genetic markers) significantly influencing the probabilities of an individual being classified into some specific categories are sought. The F2 population was categorized into resistant and susceptible based on the DI distribution gap, which was same as the segregation test. A permutation test was carried out with 5,000 replications to obtain the significance threshold of the logarithm of odds (LOD) scores.
Marker validation
Three F2 plants (F2-37, F2-73, and F2-78), that were heterozygous for the KBrS003010N1 marker (forward: 5′-TCACTCCTCTCGCAGATTCA-3′; reverse: 5′-TGGAATCGCTTTAAGCAGATGC-3′) were selected, and F3 lines were generated by self-pollination. To detect DNA polymorphisms, healthy leaves were harvested before fusarium wilt symptoms appeared. Total genomic DNA of each plant was isolated from either fresh or freeze-dried leaves using the CTAB method (Murray and Thompson 1980). Thereafter, the same procedures which were used to analyze F2 plants were used to characterize the genotypes and DIs of F3 plants. Inoculation tests were conducted in growth chambers. The F2-37 line (F3 plants) were incubated at 25°C for 7 days following transplantation, and then the temperature was changed to 28°C. Inoculation tests for the F2-78 line were maintained at a constant 28°C. Two populations of F2-73 were incubated in each of the two temperature conditions.
Comparative analysis of QTL region
To identify syntenic regions between the Foc-Bo1 region and Brassica rapa, markers designed with the bacterial artifical chromosome (BAC) sequences released from BrGP were aligned with the JWF3p map published on the BrGP web site (http://www.brassica-rapa.org/BRGP/index.jsp). The nucleotide sequences of each B. rapa BAC were aligned with Arabidopsis genome sequences by BLASTN in DDBJ (http://blast.ddbj.nig.ac.jp/). Based on a threshold value of less than e−10, the regions having conserved collinearity with A. thaliana were regarded as homologous syntenic regions.
Results
Response to fusarium resistance
In greenhouse inoculation tests, individual plant DI scores were recorded for the parents (Anju and GC), and both F1 and F3 progenies. DI scores were determined 14 days after inoculation (Fig. 1, Electronic Supplementary Material Fig. S2). Anju parental plants are typically resistant to fusarium wilt, and in our experimental protocol these plants exhibited complete resistance (DI = 0). In contrast, plants of the GC susceptible parent succumbed to the disease (DI = 2.0). F1 plants were uniformly resistant to the disease, with a mean DI of 0.73 ± 0.46. For the F2 segregation test, the DIs of individual F2 plants were deduced from inoculation test results of F3 progeny (F2:3). Based on the gap in DI distribution, plants having a DI below 1.4 (n = 66) or over 1.6 (n = 19) were classified as disease-resistant or disease-susceptible, respectively (Fig. 1). The observed segregation ratio was tested using χ2 analysis to determine whether the data fit the 3:1 segregation ratio that was expected for a single dominant gene (Table 1). The resulting χ2 value for the observed segregation ratio was 0.14 (0.7 < P < 0.8), indicating that the F2 segregation test supported the single dominant gene model.
Linkage analysis
The segregation data indicated that a single dominant gene controlled the disease-resistance trait. Based on this assumption, we analyzed DI data from 85 F2:3 plants and identified a single genetic locus for fusarium wilt resistance. This locus, which was named Foc-Bo1, was mapped to the interval between the markers BSA8 and KBrS003O10N1 on linkage group seven (O7) (Fig. 2a). The linkage map constructed by Nagaoka et al. (2010) was modified in this study by adding new markers; a total of 261 markers were distributed in nine linkage groups covering 943.7 cM, and the average interval between markers was 3.7 cM. The modified linkage map of O7 contained 28 DNA markers that included 20 simple sequence repeat (SSR), three sequence-related amplified polymorphism, two cleaved amplified polymorphic sequence, one sequence characterized amplified region and two single nucleotide polymorphism/insertion–deletion markers (Electronic Supplementary Material Table S1).
Detection of the position of fusarium wilt resistance gene in B. oleracea. a Map of B. oleracea O7 resulting from analysis of F2 population. Marker positions are indicated in centimorgans (cM) on the left side of the linkage group, and locus designations are provided on the right side. The Foc-Bo1 locus controls resistance to fusarium wilt. Distances between Foc-Bo1 and the adjacent markers are indicated in the enlarged genomic region containing Foc-Bo1. b LOD plots for QTL2 of fusarium wilt resistance by QTL analysis. LOD score and map distance are given on the x and y axes, respectively. Triangles on the x axes indicate the marker positions. Threshold of 2.01 is shown by broken line
The phenotypic DI at the KBrS003O10N1 marker closely linked to the Foc-Bo1 locus indicated that higher disease resistance was associated with the homozygous Anju genotype versus the homozygous GC genotype. Plants that were heterozygous at this locus had varying levels of resistance (Fig. 1).
QTL analysis
QTL analysis was performed using genotypic data from 94 F2 plants and DI data from 85 F2:3 plants. Because the DI distribution was a discontinuous rank with a significant gap and finite ends rather than a normal distribution, the interval mapping method for categorical traits was applied in QTL analysis. As described in “Materials and Methods”, F2 plants were divided into two categories (resistance and susceptible) based on the gap in DI distribution. We adopted an empirical value of 2.01 that corresponded to the genome-wide 1% significance level obtained with permutation tests as a threshold value for LOD scores. Two QTL for fusarium wilt resistance were identified by our analysis. QTL1 and QTL2 were mapped to O4 and O7, respectively. The major QTL (LOD 19.5) affecting fusarium wilt resistance (QTL2) was located on O7 with the proximal marker of KBrS003O10N1 (Fig. 2b). This locus was also identified by linkage analysis, which mapped fusarium wilt resistance to the Foc-Bo1 locus (30.7 cM on O7). Thus, QTL2 could be identical to Foc-Bo1. As shown in Table 2, the allele from Anju at Foc-Bo1 locus had a significant effect increasing the probability of fusarium wilt resistance. The probability of F2 individuals with the allele from Anju being classified as resistant was close to 1 (1.00 of QQ and 0.98 of Qq), while the probability of resistance was 0 (qq) for the plants without the Anju allele. Other than O7, a part of O4 showed a LOD score (2.06) slightly higher than threshold (2.01), and this locus was named QTL1. The probability of being resistant was higher for the plants with the GC allele, indicating that the resistance gene came from GC (Table 2). No other genomic region showed a LOD score higher than the threshold.
Associations between molecular markers and fusarium wilt resistance
KBrS003O10N1 was the closest marker to the gene Foc-Bo1, with only 1.2 cM between them (Fig. 2a). To validate the association between KBrS003O10N1 and Foc-Bo1, three F3 lines were selected for further analysis and tested using two temperature regimens (Fig. 3).
Frequency distribution of disease severity index (DI) for three F3 populations. The name of each population is shown above the graph, followed by the mean DI in parentheses. Experimental temperatures are indicated. F3 plants homozygous for the Anju KBrS003O10N1 locus, homozygous for the GC KBrS003O10N1 locus, or heterozygous at the KBrS003O10N1 locus are indicated by open, black, or gray bars, respectively
In F3 progeny tests at 25/28°C, when plants of each line (F2-37, F2-73) were classified either as disease resistant (DI ≤ 1) or susceptible (DI = 2), the segregation ratio of the populations fit the expected 3:1 ratio by χ2 analysis (Table 1). This indicated that a single dominant gene controlled fusarium wilt resistance, which confirmed our F2 segregation analysis. For the 28°C test, although the F2-78 line fit the 3:1 resistant:susceptible segregation ratio, the segregation of the F2-73 line deviated from 3:1, due to the increase of infection of plants heterozygous for the KBrS003O10N1 locus. The mean DI of plants heterozygous for KBrS003O10N1 locus was significantly different between the 25/28°C test and the 28°C test on the same line (F2-73) by Duncan’s new multiple range test (P < 0.01) (data not shown), indicating that the heterozygous genotypes have a lower level of resistance than the homozygous genotypes, especially at 28°C.
In F3 progeny tests, the genotype of each plant was also determined via PCR analysis (Electronic Supplementary Material Fig. S3). Most plants homozygous for the Anju KBrS003O10N1 locus were disease-resistant (DI = 0) (Fig. 3). Genotypes heterozygous for the KBrS003O10N1 locus exhibited variable levels of disease resistance, at 28°C in particular. Plants homozygous for the GC KBrS003O10N1 locus were mostly susceptible (DI = 2); however a small number of plants were either moderately resistant (DI = 1, n = 5), or completely resistant (DI = 0, n = 3). Genotype analysis further indicated that resistant plants which were homozygous for the GC genotype at KBrS003O10N1 were heterozygous at BSA8 [the locus on the other side of Foc-Bo1 (Fig. 2a)]. This indicated that a crossover event had occurred between the KBrS003O10N1 and BSA8 loci. Similar results were seen with the moderately resistant plants (three of five exhibited a crossover event). Therefore, recombinant plants that were homozygous for the GC genotype at the KBrS003O10N1 locus could be heterozygous at the Foc-Bo1 locus. This may explain why some plants that were homozygous for the GC KBrS003O10N1 locus were not susceptible to fusarium wilt, though stochastic escape from infection of susceptible plants (i.e., environmental effects or experimental errors) might have contributed to these conflicting results.
Discussion
Inheritance of fusarium wilt resistance in B. oleracea
In a cross between fusarium wilt-resistant and fusarium wilt-susceptible plants, segregation analyses indicated that a single dominant gene controlled resistance against the F. oxysporum f. sp. conglutinans Cong: 1-1 strain in B. oleracea. In addition, disease resistance was seen at high environmental temperatures (28°C). These results are consistent with Type A resistance, which has been described in cabbage (Walker 1930; Walker and Blank 1934). The gene responsible for this resistance, Foc-Bo1, was closely linked to the genetic marker KBrS003O10N1 by both linkage and QTL analyses. Foc-Bo1 was the major QTL that emerged from this analysis, of which the LOD score was 19.5. Combined with results from the marker validation test, these findings indicate that Foc-Bo1 plays a major role in fusarium wilt resistance. To our knowledge, this is the first report that has successfully mapped a fusarium-resistance gene and verified the linkage relationship between the resistance gene and the closest genetic marker in Brassica species.
Blank (1937) reported that Wisconsin All Seasons cabbage cultivar carried the two types of resistance, Type A and Type B. With the expectation that fusarium resistance is a multiple trait in B. oleracea, QTL analysis was applied to an F2 population. Other than Foc-Bo1, a minor QTL, QTL1, was detected on O4. Although the LOD value of 2.06 on O4 was slightly higher than the 2.01 threshold, further analysis is required to determine whether QTL1 represents a legitimate locus involved in fusarium wilt resistance.
Temperature effects on fusarium wilt resistance
Temperature plays an important role in fusarium wilt resistance. Type B resistance is polygenic and becomes phenotypically unstable at temperatures above 24°C. In contrast, Type A resistance is stable at 26–28°C (Walker and Smith 1930; Blank 1937; Walker 1953). Whether Type A resistance becomes unstable above 28°C is unclear. Walker and Smith (1930) reported that for Type A resistance, a small number of diseased plants appear at 26°C, and disease incidence increases as the temperature rises to 28°C. At 26–28°C, the fusarium wilt symptoms in Type A-resistant cabbage are not typical of yellows. Instead, the plants become stunted, and exhibit a pale yellowing of the leaf parenchyma. In contrast, Farnham et al. (2001) observed no significant differences between Type A resistant cabbage (Bravo) under 25/25°C (day/night) and 30/25°C conditions. In our current study, plants homozygous for the resistant Anju genotype were stably resistant in both 25/28°C and constant 28°C growth chamber conditions, although a few Anju genotypes were infected at 28°C in the F3-73 line. Discrepancies between research groups may have resulted from different growth chamber conditions. Walker and Smith (1930) performed experiments in which the soil temperature in the incubation tank (26–28°C) was higher than the temperature of the air (15–17°C). In our study and Farnham et al. (2001), the soil and air temperatures were equivalent.
In this study, we identified plants that were heterozygous at the Foc-Bo1 locus. Plants having only one copy of the disease-resistance allele were partially resistant to fusarium wilt and were more disease-susceptible at high temperatures. Foc-Bo1, therefore, acts in a dose-dependent manner to control fusarium wilt. This result is consistent with Walker and Smith (1930), who crossed resistant and susceptible parents to generate heterozygous progeny. These heterozygotes were disease-resistant at 24°C, but became stunted and yellow at 26°C. In contrast, homozygous resistant plants were quite healthy at 26°C. Stable resistance to clubroot disease also requires that clubroot-resistance genes are homozygous (Suwabe et al. 2003; Nomura et al. 2005; Nagaoka et al. 2010). Factors that regulate the cumulative effects of disease-resistance genes have not been identified.
Marker validation for MAS
In this study, we used the SSR marker KBrS003O10N1 to track the disease-resistance allele of Foc-Bo1. This method is both technically simple and inexpensive (Collard et al. 2005). During the F3 progeny test (Fig. 3), KBrS003O10N1 proved extremely effective in identifying the fusarium wilt-resistance genotype in B. oleracea. This method was not always accurate, however, as recombination events were detected between KBrS003O10N1 and BSA8, the markers that flank Foc-Bo1. As recombination between the KBrS003O10N1 and Foc-Bo1 loci is clearly possible, genotyping using the two markers (KBrS003O10N1 and BSA8) would more accurately identify plants with the disease-resistance allele of Foc-Bo1. BSA8 is 4.6 cM away from Foc-Bo1, however, which allows for a high rate of recombination between these two loci. Therefore, to increase the accuracy of MAS, a novel marker that is more closely linked to Foc-Bo1 must be generated to replace BSA8. Recently, Brassica genomics resources have been disclosed (http://www.brassica.info/; http://www.brassica-rapa.org/BRGP/geneticMap.jsp), and will facilitate fine mapping of the FocBo1 region.
The artificial inoculation test could not distinguish between plants that were heterozygous or homozygous at the Foc-Bo1 locus. As such, progeny tests were required to accurately assess the Foc-Bo1 genotype, which were quite time-consuming. In contrast, using markers linked to the Foc-Bo1 locus to identify heterozygous plants was technically practical and inexpensive. Moreover, KBrS003O10N1 analysis identified plants that carried the resistance allele in the F2 population. This is important because artificial inoculation tests on single plants within the F2 population were not reliable.
Race/species-specific resistance gene
Although F. oxysporum f. sp. conglutinans is the primary pathogen that causes fusarium wilt in B. oleracea, F. oxysporum f. sp. raphani can also infect B. oleracea (Garibaldi et al. 2006). In addition, Type A resistance in cabbage is race-specific; these plants are resistant to f. sp. conglutinans race 1, but can be infected by race 2 (Ramirez-Villupadua et al. 1985; Morrison et al. 1994). In Arabidopsis thaliana, RFO1 (on chromosome 1) is non-race-specific, conferring resistance to f. sp. matthioli, f. sp. conglutinans, and f. sp. raphani, which are causal agents of fusarium wilt in Brassicaceae (Diener and Ausubel 2005). The syntenic regions in A. thaliana and B. rapa, corresponding to the Foc-Bo1 region, are illustrated in Electronic Supplementary Material Fig. S4, based on the result of Nagaoka et al. (2010). In this study, we revised the marker order to perform precise syntenic analysis. As a result, we confirmed that the central region of B. oleracea O7 was aligned with the corresponding region of chromosomes 3 and 4 of A. thaliana but not with chromosome 1 of A. thaliana. Besides, only one polymorphic band of a RFO1 homologue was detected in GC and Anju. BoRFO1 was then mapped to O2 in our map (data not shown). We therefore conclude that RFO1 (on At chr1) is not a candidate gene for Foc-Bo1. Thus, it will be interesting to learn the reaction of Foc-Bo1 against the related formae speciales and races, as well as the mechanism by which Foc-Bo1 overcomes fusarium wilt.
This study showed that the central region of O7 in B. oleracea contains Foc-Bo1. Nagaoka et al. (2010) reported that this region contains the clubroot-resistance gene, Pb-BoAnju4 (Electronic Supplementary Material Fig. S4). In addition, Nagaoka et al. (2010) suggested that the central region of O7 in B. oleracea might also correspond to the region harboring the clubroot-resistance gene (CRb) in R3 of B. rapa and chromosome 4 of A. thaliana that contains a disease-resistance gene cluster (Mayer et al. 1999; Young 2000). In this way, the central region of O7 of B. oleracea contains multiple disease-resistance genes, Pb-BoAnju4 and Foc-Bo1 at least, indicating the importance of this genomic region to the B. oleracea breeding program. Molecular cloning of Foc-Bo1 and extensive analysis of the surrounding genomic region will be necessary to understand the molecular and evolutionary bases of disease resistance in Brassica.
References
Anderson EM (1933) Fusarium resistance in Wisconsin Hollander cabbage. J Agric Res 47:639–661
Berrocal-Lobo M, Molina A (2007) Arabidopsis defense response against Fusarium oxysporum. Trends Plant Sci 13:145–150
Blank LM (1937) Fusarium resistance in Wisconsin all seasons cabbage. J Agric Res 55:407–510
Bosland PW, Williams PH, Morrison RH (1988) Influence of soil temperature on the expression of yellows and wilt of crucifers by Fusarium oxysporum. Plant Dis 72:777–780
Collard BCY, Jahufer MZZ, Brouwer JB, Pang ECK (2005) An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: the basic concepts. Euphytica 142:169–196
Diener AC, Ausubel FM (2005) RESISTANCE TO FUSARIUM OXYSPORUM 1, a dominant Arabidopsis disease-resistance gene, is not race specific. Genetics 171:305–321
Dixon GR (2007) Vegetable brassicas and related crucifers. CABI Publishing, Wallingford
Farnham MW, Keinath AP, Smith JP (2001) Characterization of fusarium yellows resistance in collard. Plant Dis 85:890–894
Garibaldi A, Gilardi G, Gullino ML (2006) Evidence for an expanded host range of Fusarium oxysporum f. sp. raphani. Phytoparasitica 34:115–121
Hayashi T, Awata T (2006) Interval mapping for loci affecting unordered categorical traits. Heredity 96:185–194
Iwata H, Ninomiya S (2006) AntMap: constructing genetic linkage maps using an ant colony optimization algorithm. Breed Sci 56:371–377
Kikuchi S, Takata S, Ichii M, Sawasaki S (2003) Efficient fine mapping of the naked caryopsis gene (nud) by HEGS (High Efficiency Genome Scanning) AFLP in barley. Theor Appl Genet 108:73–78
Mayer K, Schuller C, Wambutt R, Murphy G, Volkaert G, Pohl T, Dusterhoft A, Stickema W, Entian K-D, Terryn N et al (1999) Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature 402:769–777
Morrison RH, Mengistu A, Williams PH (1994) First report of race 2 of cabbage yellows caused by Fusarium oxysporum f. sp. conglutinans in Texas. Plant Dis 78:614
Murray M, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Nagaoka T, Doullah MAU, Matsumoto S, Kawasaki S, Ishikawa T, Hori H, Okazaki K (2010) Identification of QTLs that control clubroot resistance in Brassica oleracea and comparative analysis of clubroot resistance genes between B. rapa and B. oleracea. Theor Appl Genet 120:1335–1346
Nomura K, Minegishi Y, Kimizuka-Takagi C, Fujioka T, Moriguchi K, Shishido R, Ikehashi H (2005) Evaluation of F2 and F3 plants introgressed with QTLs for clubroot resistance in cabbage developed by using SCAR markers. Plant Breed 124:371–375
Ramirez-Villupadua J, Endo RM, Bosland P, Wiliams PH (1985) A new race of Fusarium oxysporum f. sp. conglutinans that attacks cabbage with type A resistance. Plant Dis 69:612–613
Snyder WC, Hansen HN (1940) The species concept in Fusarium. Am J Bot 27:64–67
Suwabe K, Tsukazaki H, Iketani H, Hatakeyama K, Fujimura M, Nunome T, Fukuoka H, Matsumoto S, Hirai M (2003) Identification of two loci for resistance to clubroot (Plasmodiophora brassicae Woronin) in Brassica rapa L. Theor Appl Genet 107:997–1002
Walker JC (1930) Inheritance of fusarium resistance in cabbage. J Agric Res 40:721–745
Walker JC (1953) Cauliflower, cabbage, and others. In: Stefferud A (ed) Plant diseases. US Dept Agric, Washington, DC, pp 425–430
Walker JC, Blank LM (1934) Fusarium resistant Danish Ballhead cabbage. J Agric Res 49:983–989
Walker JC, Smith R (1930) Effect of environmental factors upon the resistance of cabbage to yellows. J Agric Res 41:1–15
Walker JC, Monteith J Jr, Wellman FL (1927) Development of three mid-season varieties of cabbage resistant to yellows (Fusarium conglutinans Woll.). J Agric Res 35:785–810
Young ND (2000) The genetic architecture of resistance. Curr Opin Plant Biol 3:285–290
Acknowledgments
The authors sincerely thank Dr. Kadota at the National Agricultural Research Center for Tohoku Region, Japan, for kindly providing the F. oxysporum f. sp. conglutinans strain. This work was supported in part by a grant from Japan Science and Technology Agency, Japan (Adaptable and Seamless Technology Transfer Program through Target-Driven R&D No. AS221Z02511).
Author information
Authors and Affiliations
Corresponding author
Additional information
Zi-jing Pu and Motoki Shimizu contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pu, Zj., Shimizu, M., Zhang, Yj. et al. Genetic mapping of a fusarium wilt resistance gene in Brassica oleracea . Mol Breeding 30, 809–818 (2012). https://doi.org/10.1007/s11032-011-9665-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-011-9665-8