Abstract
Pollen fertility restoration of the CMS phenotype caused by H. chilense cytoplasm in wheat was associated with the addition of chromosome 6HchS from H. chilense accession H1. In order to develop an euploid restored line, different genomic combinations substituting the 6HchS arm for another homoeologous chromosome in wheat were evaluated, with the conclusion that the optimal combination was the translocation T6HchS·6DL. The double translocation T6HchS·6DL in H. chilense cytoplasm was obtained. This line is fertile and stable under different environmental conditions. However, a single dose of the T6HchS·6DL translocation is insufficient for fertility restoration when chromosome 6D is also present. Restoration in the msH1 system is promoted by interaction between two or more genes, and in addition to the restorer of fertility (Rf) located on chromosome 6HchS, one or more inhibitor of fertility (Fi) genes may be present in chromosome 6DL.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Heterosis is a powerful tool for improving yield and quality in many crops. Hybrid rice and hybrid maize have contributed to enhanced productivity, which is essential to supply enough food for an increasing world population. The recent success of hybrid rice in China has led to continuing interest in hybrid wheat, even when most research in this area has been discontinued in many countries for reasons including low heterosis and high seed production costs. To date no cytoplasmic male sterility (CMS) system giving stable sterility with no deleterious side-effects from the cytoplasm and with a high degree of restoration has been obtained in wheat. The two CMS systems which are currently most widely used, the G-type system (Triticum timopheevi cytoplasm) and the Sv-type system (Aegilops kotschyi cytoplasm) have clear disadvantages, including unstable sterility and morphological disturbances (Wilson and Driscoll 1983; Tsunewaki 1993). Nevertheless, the development of commercial hybrid wheat remains of great interest, and a key to success will be to improve hybrid production efficiency, for which finding better sources of male sterile cytoplasm is a critical requirement. A new CMS source in bread wheat (Triticum aestivum L.) designated msH1 has recently been reported (Martín et al. 2008a). This system uses the cytoplasm of Hordeum chilense Roem. et Schult (2n = 2x = 14, HchHch), a diploid wild barley native to Chile and Argentina which posses some traits potentially useful for wheat breeding (Martín et al. 1996; Atienza et al. 2004, 2007a, b) and which exhibits high crossability with others members of the Triticeae tribe (Bothmer and Jacobsen 1986; Martín et al. 1998). The male sterility of alloplasmic wheat containing H. chilense cytoplasm is stable under different environmental conditions and it does not exhibit developmental or floral abnormalities, showing only slightly reduced height and some delay in heading. Considering the features displayed by this system, it offers a real potential for the development of viable technology for hybrid wheat production. Fertility restoration of the CMS phenotype caused by the H. chilense cytoplasm has been associated with the addition of the short arm of chromosome 6Hch (6HchS) from H. chilense accession H1. Thus, some fertility restoration genes appear to be located on this chromosome arm. The fertility of the restored line T593 (disomic addition of 6HchS to wheat in H1 cytoplasm) was seen to be stable but lower than euplasmic wheat (Martín et al. 2008a). It is known that aneuploidy frequently causes a decrease in fertility and therefore, the lower fertility of T593 when compared with euplasmic wheat may be due in part to this effect and not only to the H1 cytoplasm. Indeed, experimental data show that a ditelosomic addition of 6HchS in wheat line T21 presents 12% less grain yield than T21 when grown under the same conditions (Martín et al. 2008a). Moreover, the possibilities of using H. chilense cytoplasm for hybrid seed production are null if the system is based on restoration by aneuploidy. Therefore, the development of an euploid restored line is absolutely necessary.
At present, most donor species of restoring genes from other CMS systems come from the genus Triticum and their genes can be transferred to wheat easily. These restoring genes were introduced into wheat chromosomes mainly through homologous crossing-over which occurs naturally (Wilson 1984). Transfer of alien genes from more distant species to wheat can be accomplished by irradiation, genetic manipulation or the homoeologous pairing between wheat and alien chromosomes and selection of spontaneous translocations including centromeric translocation. The centric breakage-fusion behaviour of univalents can be exploited to transfer whole alien chromosome arms to wheat (Sears 1952). The alien chromosome and a homoeologous wheat chromosome are isolated in monosomic condition. In such double monosomic plants, both monosomes stay as univalent at meiotic metaphase I. Univalents have a tendency to break at the centromeres, followed by fusion of the broken arms, giving rise to Robertsonian whole arm translocations (Robertson 1916). Depending on the chromosomes involved and the environmental conditions, the desired compensating wheat-alien Robertsonian translocations can be recovered at frequencies, ranging from low to almost 20% (Davies et al. 1985, Lukaszewski and Curtis 1993; Lukaszewski 1994, 1997).
However, fertility restoration is usually a complex genetic trait. Most workers have concluded that the inheritance of male fertility restoration is controlled by 2 or 3 major Rf genes and modifiers located on 17 of the 21 wheat chromosomes (Sage 1976; Maan et al. 1984) and some authors point out that genes on all 21 chromosomes may be involved in cytoplasmic male sterility and fertility restoration in wheat (Du et al. 1991).
Therefore, in addition to obtaining the translocation of the 6HchS chromosome arm into the wheat genome, our aim is to obtain different genomic combinations substituting the 6HchS arm for other homoeologous chromosomes in wheat, with the aim of establishing the optimal translocation (6HchS·6AL, 6HchS·6BL or 6HchS·6DL). In our experience, we observe that substitution of group B chromosomes is less well tolerated than A and D chromosomes. Therefore, considering the great amount of crossing work required by this study, we focused on chromosomes 6A and 6D.
The present work was aimed at obtaining the most favourable translocation of the 6HchS chromosome arm into the wheat genome to avoid the effect caused by aneuploidy and at evaluating the fertility and morphological characteristics of the lines.
Materials and methods
Plant material
The genetic stocks used in this study are shown in Table 1. Lines T21A6H1, T529 and T530 were kindly provided by Steve Reader, JIC, Norwich, UK. Lines T218 and T593 are described in Martín et al. (2008a).
Several crosses were carried out using this plant material during five consecutive years in order to obtain different genomic combinations involving the 6Hch and 6HchS chromosomes in an alloplasmic wheat background with H. chilense cytoplasm. All plants were grown in the greenhouse. The temperature was maintained at 7–25°C (night/day), and light was supplemented to maintain a 13-h photoperiod. During the heading stage, the spikes were bagged, both for selfing or crossing as female parent.
Cytological observations
For somatic chromosome counting, root tips 1-cm long were collected from germinating seeds and pre-treated for 4 h in an aqueous colchicine solution (0.05%) at 25°C. They were fixed in a freshly prepared 3 absolute ethanol: 1 glacial acetic acid (v/v) mixture and stained by the conventional Feulgen technique.
For meiotic chromosomes observation, anthers were collected and stained directly with 0.1% acetocarmine.
Molecular analysis
DNA was extracted from 5 to 6-week-old T218, T593, T21, T21A6H1, T529, T530 and H. chilense (H1) seedlings according to the procedure of Doyle and Doyle (1990) with minor modifications. The chloroplast consensus simple sequence repeat ccSSR-4 developed by Chung and Staub (2003) was used to verify the presence of the H. chilense cytoplasm in the alloplasmic lines (Atienza et al. 2007c; Martín et al. 2008b). The polymerase chain reaction (PCR) was carried out in 25 μl of reaction mixture as described by Chung and Staub (2003).
The expressed sequence tag (EST) markers k01062 and k03014 (Hagras et al. 2005; Nasuda et al. 2005) were used to identify the 6HchS and the 6HchL chromosomes respectively. The marker k01062 amplifies differently with PCR in bread wheat and barley (H. vulgare cv. Betzes), and it is assigned to the short arm of chromosome 6H in barley. Moreover, it was also shown to be amplified in chromosome 6Hch in H. chilense (Martín et al. 2008a). PCR was carried out as described by Nasuda et al. (2005).
The microsatellite simple sequence repeats markers Xgdm 127 and Xgdm 98 (Röder et al. 1998; Pestsova et al. 2000) were used to identify the 6DS and 6DL respectively. PCR was carried out as described by Röder et al. (1998). All amplification products were resolved by agarose gel electrophoresis and visualised with ethidium bromide.
Fluorescence in situ hybridization (FISH)
Root tips were fixed as described above. Preparations were made as described by Prieto et al. (2001).
Probe pTa71, containing 1 unit of 18S-5.8S-26S rDNA (8.9 kb) from T. aestivum (Gerlach and Bedbrook 1979) was labelled by nick translation with biotin-11-dUTP (Roche Corporate, Basel, Switzerland) and total T21 DNA was labelled with digoxigenin-dUTP. Both probes were mixed in the hybridization solution to the final concentration of 5 ng/μl. After examination of nuclei hybridized with the repetitive DNA probe and genomic T21 DNA, preparations were re-probed using the total genomic DNA of H. chilense as probe. Total H. chilense DNA was labelled by nick translation with biotin-11-dUTP. The in situ hybridization protocol was that of Cabrera et al. (2002). Digoxigenin- and biotin-labelled probes were detected with antidigoxigenin-FITC (Roche Corporate) and streptavidin-Cy3 conjugates (Sigma, St. Louis, MO, USA), respectively. Chromosomes were counterstained with DAPI (40,6-diamidino-2-phenylindole) 339 and mounted in Vectashield (Vector Laboratories Inc.). Slices were examined by using a Zeiss LSM 5 Pascal confocal laser scanning microscope with LSM 5 Pascal software version 3.0 (Zeiss, Jena, Germany), and processed with PhotoShop 7.0 software (Adobe Systems Inc., San Jose, CA, USA).
Open field experiments
A completely randomised block design with 5 replications was conducted under field conditions, to evaluate the morphology and fertility of T21 (T. aestivum cv. Chinese Spring), T218 (T21 in H. chilense cytoplasm) and two newly-obtained lines: the Chinese Spring disomic substitution 6Hch(6D) in H. chilense cytoplasm named T635, and the double translocation T6HchS·6DL in H. chilense cytoplasm named T650. Five plants of each genotype were grown in the I.A.S. in Córdoba (Spain) from February to June. Data of plant height, tillers per plant, spikelets per spike and seed per plant were collected, and analysis of variance was carried out using the least significant difference method (l.s.d.) method (P ≤ 0.05). Statistical analyses were performed with Statistix v. 8.0.
Results
Crossing scheme and fertility scoring
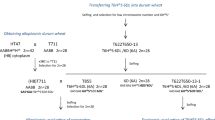
Figures 1 and 2 show the breeding procedure carried out in this work to obtain the different genomic combinations involving the 6HchS and 6Hch chromosomes in the alloplasmic wheat background with H. chilense cytoplasm. All genomic combinations display normal female fertility but different degrees of male sterility. In order to facilitate the reading of the breeding procedure described in Figs. 1 and 2, the genetic combinations obtained are detailed in Tables 2 and 3 respectively.
Crossing scheme showing the different genomic combinations obtained involving the 6HchS chromosome in alloplasmic wheat background with H. chilense cytoplasm. Only (H1)CS-Hch M6D-DtA6HchS (20″+1′6D+t″6HchS) displays normal pollen fertility. T593: (H1)CS-Hch DtA6HchS (21″+t″6HchS); T529: CS N6D (20″+0″6D); T530: CS N6A (20″+0″6A)
Breeding procedure to obtain the different genomic combinations involving the 6Hch chromosome in alloplasmic wheat background with H. chilense cytoplasm. Only (H1)CS-Hch M6D-M6Hch (20″+1′6D+1′6Hch) and (H1)CS-Hch DS6Hch(6D) (20″+1″6Hch(6D)) present normal pollen fertility. T21A6H1: CS-Hch DA6Hch (21″+1″6Hch); T529: CS N6D (20″+0″6D); T530: CS N6A (20″+0″6A)
In Fig. 1, T593 ((H1)21″+t″6HchS) was emasculated and pollinated with T529 (20″+0″6D) (Fig. 1a) and T530 (20″+0″6A) (Fig. 1b). The progenies were completely male sterile and were pollinated with T593. The genomic composition of the progenies was determined cytologically by somatic chromosome counting. As shown in Fig. 1a, the genotypes (H1)21″+t′6HchS and (H1)20″+t″6HchS(6D) displayed some degree of male fertility although it was extremely low. However, (H1)20″+1′6D+t″6HchS presented normal male fertility. On the other hand, when chromosome 6A was absent (Fig. 1b), neither of the genomic combinations displayed a high degree of fertility.
Crosses involving the 6Hch chromosome in the alloplasmic wheat background are detailed in Fig. 2. Briefly, alloplasmic line T218 was pollinated with T21A6H1 (21″+1″6Hch). Progeny was analyzed cytologically and, as expected, the genomic configuration was (H1)21″+1′6Hch. Seeds were grown and the plants resulted male sterile, so they were backcrossed to T529 and T530. Their progenies were again screened cytologically by somatic chromosome counting. Both (H1)20″+1′6A and (H1)20″+1′6D were male sterile. However, while (H1)20″+1′6A+1′6Hch was also male sterile (H1)20″+1′6D+1′6Hch was completely fertile.
The main goal of this work was to obtain the most favourable translocation of the H. chilense 6HchS chromosome arm into the wheat genome by the centric breakage-fusion behaviour of univalents. Our results suggest that substitution of 6Hch for 6D is superior to substitution for 6A. For that reason, we focused on obtaining genomic combinations involving chromosomes 6D and 6Hch. The genomic combination (H1)20″+1′6D+1′6Hch resulted completely fertile, therefore, we decided to engineer the translocation T6HchS·6DL by consecutive self-fertilization of this genotype.
Selection by molecular markers and FISH of the T6HchS·6DL translocation
Self-progenies of (H1)20″+1′6D+1′6Hch were analysed cytologically at mitosis and by molecular markers to identify the T6HchS·6DL translocation. EST markers k01062 and k03014 were used to identify the 6HchS and the 6HchL chromosome respectively. The microsatellite simple sequence repeat markers Xgdm 127 and Xgdm 98 were used to identify 6DS and 6DL respectively. All amplification products were resolved by agarose gel electrophoresis and visualised with ethidium bromide.
Markers k01062 and Xgdm 98 were amplified for the purpose of the present work in self-progeny of (H1)20″+1′6D+1′6Hch. Those plants which displayed amplification products from both molecular markers were selected. Next, fluorescence in situ hybridization (FISH) was carried out on this plant material to investigate if the T6HchS·6DL translocation was present. Genomic in situ hybridization (GISH) using H. chilense H1 genomic DNA as probe was used to identify the 6HchS chromosome. This chromosome arm was present in all plants as indicated by molecular marker. In some of them, it was present the whole 6Hch chromosome and in some others only the telosomic arm 6HchS. However, it was not possible to identify the T6HchS·6DL translocation.
Genomic combination (H1)20″+1′6D+1′6Hch was selected again from this material and selfed twice until the translocation T6HchS·6DL was obtained (Fig. 3a). Although the marker k01062 was sufficient to confirm the identity of the H. chilense chromosome arm, FISH was also carried out on this plant using the pTa71 probe (Fig. 3b). This probe contains 1 unit of 18S-5.8S-26S rDNA (located in the nucleolar organiser regions (NORs) of chromosomes). The NORs in Hordeum are localised in chromosome segments 5HS and 6HS, therefore, the presence of the pTa71 probe signal in the H. chilense chromosome, further confirms the results of molecular marker assays in identifying the presence of 6HchS chromosome arm.
In situ hybridization to root-tip metaphase cells from monosomic translocation T6HchS·6DL in H1 cytoplasm. a GISH using H. chilense H1 genomic DNA probe (detected with streptavidin-Cy3, red). Blue DAPI staining shows the wheat chromosomal DNA. b Double FISH signals using the pTa71 probe (detected with streptavidin-Cy3, red) and T21 genomic DNA probe (detected with antidigoxigenin-FITC, green). Wheat chromosomes show an intense green colour while 6HchS chromosome arm only displays the red pTa71 probe signal (indicated by an arrow). Satellited wheat chromosomes also show the pTa71 signal
The plant with the T6HchS·6DL translocation was male fertile, but in order to use it as a restorer line, it is necessary to develop this translocation in disomic condition. Therefore, the plant was again selfed, and progeny was screened to identify the (H1)T21 double translocation T6HchS·6DL. Progenies were first analysed cytologically at mitosis, selecting for chromosome number 42 and subsequently sown. Next, this material was analyzed by molecular markers. Those genotypes displaying amplification products only with k01062 and Xgdm 98 markers (but no amplification with k03014 and Xgdm 127) were selected. This procedure should identify plants with the genotype (H1)T21 double translocation T6HchS·6DL.
To confirm this result, GISH using H. chilense H1 genomic DNA as probe was performed and the double translocation T6HchS·6DL was verified (Fig. 4a). This line was fertile and was designated T650: (H1)20″+1″T6HchS·6DL.
Genomic in situ hybridization to root-tip metaphase cells using H. chilense H1 genomic DNA probe (detected with streptavidin-Cy3, red) and T21 genomic DNA probe (detected with antidigoxigenin-FITC, green) from: a Double translocation T6Hch·6DL in H1 cytoplasm (named T650). b Disomic substitution 6Hch(6D) in H1 cytoplasm (named T635). Wheat chromosomes show green colour while 6Hch and 6HchS chromosome arm are stained in red
As can be observed in Fig. 2, during the process of obtaining the T650 line, different new combinations involving chromosome 6Hch and 6D were obtained: (H1)20″+0″6D that was male sterile, (H1)20″+t″6HchS(6D) that resulted only partially fertile and (H1)20″+1″6Hch(6D) that was stable and fertile and named T635. GISH on this line using H. chilense H1 genomic DNA as probe is shown in Fig. 4b.
Finally, cytoplasm identity must be always assessed when working with alloplasmic lines. In order to prove that there was no transmission of paternal cytoplasm in the lines T650 and T635, the primer ccSSR-4 was used. This chloroplast marker is polymorphic with amplification products of 200 bp in wheat and 225 bp in H1 cytoplasm (Martín et al. 2008b). DNA of line T650 and T635 was amplified by this primer pair, confirming their alloplasmic condition.
Morphological characterisation of the double translocation T6HchS·6DL
T635 and T650 were stable under different environmental conditions: growth chamber, greenhouse and in open field (Table 4). When T635 and T650 were compared with T21 in greenhouse conditions, no difference was observed among them, except for slightly reduced height in the new combinations (data not shown). The open field is generally the most sensitive and critical environment and where differences can be detected most reliably. For that reason, a trial was conducted under field conditions to compare lines T21, T218, T635 and T650 in terms of morphology and fertility.
As seen under growth chamber and greenhouse conditions, alloplasmic lines in H. chilense cytoplasm are shorter in height than T21. The number of tillers per plant was lower in T635 and T650 than in T21 and T218, while the number of spikelets per spike was not significantly different among the four genotypes. The degree of male fertility was slightly lower in T635 than in T21; however, no significant differences were found between T21 and T650.
The final purpose of obtaining the T650 line was to use it as a restorer line to restore male fertility in the msH1 system. Therefore, the male sterile line T218 was pollinated with T650 to check if male fertility was restored. Some production of mature pollen was observed in the hybrids, but, no seed set was accomplished. T650 was also pollinated with different T. aestivum varieties: Balivial, Bandoli, Bobwhite, Pandora and T26 (a Spanish cultivar). In all the cases studied, similar results were observed (i.e. no seed set). On the other hand, when T635 and T650 were crossed, the progeny (20″+1′T6HchS·6DL+1′6Hch) displayed complete and stable fertility.
Discussion
Disomic addition of the chromosome arm 6HchS in the male sterile wheat line T218 restores completely male fertility, indicating that at least one Rf gene is present in this chromosome arm. Development of an euploid restored line is absolutely necessary for use in a restorer system, therefore, a major objective of the present research was to manipulate homoeologous group 6 chromosomes in bread wheat and H. chilense to obtain the most favourable translocation of the 6HchS chromosome arm into the wheat genome. Following the breeding procedures accomplished in this work (Figs. 1 and 2), it was concluded that the optimal combination was the translocation T6HchS·6DL. This result supports the observations already made by some authors about the similarity between the D and Hch genomes (Padilla and Martín 1983; Thomas and Pickering 1985; Cabrera et al. 1995).
However, fertility restoration is usually a complex genetic trait. Interspecific CMS and male fertility restoration systems in hexaploid wheat are conditioned by interactions involving dosage of the male fertility-restoring (Rf) and male fertility-inhibiting (Fi) genes in the polyploid nucleus, and also cytoplasmic genes (Maan and Lucken 1967; Du et al. 1991; Maan 1992). The msH1 system is not an exception. Perhaps the best example among all the homoeologous group 6 combinations is the genotype (H1)CS-Hch MA6Hch (21″+1′6Hch) vs the genotype (H1)CS-Hch M6D-MA6Hch (20″+1′6D+1′6Hch). The first combination is male sterile, while the second one is male fertile. However, meiosis of (H1)CS-Hch MA6Hch is expected to be more regular because only well-balance gametes or those with a 6Hch addition are produced. On the other hand, meiosis of (H1)CS-Hch M6D-MA6Hch can lead to many types of gametes: nulisomic 6D, euploids, 6Hch addition and different combinations involving telosomes after misdivision of univalent 6D and 6Hch, or translocations as the one obtained in this work. Therefore, restoration in the msH1 system must be promoted by interaction between two or more genes, and apart from the Rf gene located on the 6HchS, one or more inhibitor of fertility (Fi) genes may be present on chromosome 6D, specifically on the 6DL arm. Based on these results, we suggest the presence of a restorer gene on 6HchS that we will name \(Rf_{6H^{ch} S}\) and an inhibitor of fertility in chromosome 6DL that we will call Fi 6DL .
When comparing male fertility among all the combinations obtained, it can be concluded that \(Rf_{6H^{ch} S}\) and Fi 6DL dosage is a key factor in determining male fertility. The fertility of all the genomic combinations obtained in this work fit with this hypothesis. These Fi genes must be broadly extended and present most probably in most varieties of T. aestivum; at least in T21, T26, Balivial, Bandoli, Bobwhite and Pandora varieties, which were tested in this study. A pollen killer (Ki) gene has been described on chromosome 6BL in wheat and on chromosome 6SL in Aegilops (McIntosh et al. 2003). Considering the high degree of synteny among the Triticeae, these Ki genes may be homoeologous to Fi 6DL . Börner et al. (1998) suggest that some genes controlling CMS restoration are conserved across the cereal species rye and wheat, and this conservation may extend across other members of the Triticeae.
Besides Rf and Fi interactions, in G-type cytoplasm, many experiments have shown that restorer gene expression is influenced by genetic background and the interaction of genotype with environment, and that restoring genes can not completely and stably restore the fertility of G-type sterility (Tsunewaki 1980; Maan et al. 1984; Du and Maan 1992; Ikeguchi et al. 1999). In the msH1 system, stable restoration can be accomplished in T593, T650 and T635 lines; however, in genomic combinations where only partial restoration is obtained, fertility always displays an environmental response.
In our proposed CMS system, three lines would be required: a male sterile line (A-line) carrying common wheat nucleus and H. chilense cytoplasm (T218), a maintainer line (B-line) with common wheat nucleus and cytoplasm (T21) and a restorer line (R-line) that carries Rf genes with H. chilense cytoplasm. However, male fertility is not restored in the hybrid between T218 (line A) and T650. This result indicates that a single dose of the T6HchS·6DL is insufficient for stable fertility restoration when chromosome 6D is also present, meaning that T650 can not be used directly as a restorer line (R-line).
Although more than 70 different kinds of male sterile cytoplasm in common wheat have been discovered (Chen and Zhang 1994; Murai 2001, 2002; Zhang and Zhang 2001; Zhang et al. 2001; Hattori et al. 2002), the current systems of hybrid wheat production based on CMS are all of the A-line/R-line type and all share similar problems in hybrid fertility restoration. In order to restore CMS effectively, it is important that many Rf genes are accumulated into the R-line while Fi genes must be avoided in both A and R lines, which is a difficult task. Twenty-five restorer lines of T. timopheevi have already been tested by Martín et al. (2008a) in alloplasmic line T218, but no restoration was obtained; therefore, no major genes that restore the msH1 system are present in these lines. However, the G-type system displays a similar behaviour to the msH1 system, and for that reason, some other useful genes, as modifiers or fertility-conditioning genes, could be present in these restorer lines and may be of great value in terms of fertility restoration when added to the T650 line. In our future work, different cultivars will be tested with the aim of identifying Fi 6DL -deficient cultivars as well as new Rf genes that could produce self-fertile F1 hybrids.
References
Atienza SG, Ramírez MC, Hernández P, Martín A (2004) Chromosomal location of genes for carotenoid pigments in Hordeum chilense. Plant Breed 123:303–304. doi:10.1111/j.1439-0523.2004.00918.x
Atienza SG, Avila CM, Martín A (2007a) The development of a PCR-based marker for PSY1 from Hordeum chilense, a candidate gene for carotenoid content accumulation in tritordeum seeds. Aust J Agric Res 58(8):767–773. doi:10.1071/AR06338
Atienza SG, Ballesteros J, Martín A, Hornero-Mández D (2007b) Genetic variability of carotenoid concentration and degree of esterification among tritordeum (Tritordeum Ascherson et Graebner) and durum wheat accessions. J Agric Food Chem 55:4244–4251. doi:10.1021/jf070342p
Atienza SG, Martín AC, Ramírez MC, Martín A, Ballesteros J (2007c) Effects of Hordeum chilense cytoplasm on agronomic traits in common wheat. Plant Breed 126:5–8. doi:10.1111/j.1439-0523.2007.01319.x
Börner A, Korzum V, Polley A, Malyshew S, Melz G (1998) Genetics and molecular mapping of a male fertility restoration locus (Rfg1) in rye (Secale cereale L.). Theor Appl Genet 97:99–102. doi:10.1007/s001220050871
Bothmer RV, Jacobsen N (1986) Interspecific crosses in Hordeum (Poaceae). Plant Syst Evol 153:49–64. doi:10.1007/BF00989417
Cabrera A, Friebe B, Jiang J, Gill BS (1995) Characterization of Hordeum chilense chromosomes by C-banding and in situ hybridization using highly repeated DNA probes. Genome 38:435–442
Cabrera A, Martín A, Barro F (2002) In situ comparative mapping (ISCM) of Glu-1 loci in Triticum and Hordeum. Chromosome Res 10(1):49–54. doi:10.1023/A:1014270227360
Chen QF, Zhang QQ (1994) Improvement of Q-type cytoplasmic male-sterile lines and their restorers. Seeds 1:3–5
Chung S-M, Staub JE (2003) The development and evaluation of consensus chloroplast primer pairs that possess highly variable sequence regions in a diverse array of plant taxa. Theor Appl Genet 107:757–767. doi:10.1007/s00122-003-1311-3
Davies PA, Pallotta MA, Driscoll CJ (1985) Centric fusion between nonhomologous rye chromosomes in wheat. Can J Genet Cytol 27:627–632
Doyle JJ, Doyle JH (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Du H, Maan SS (1992) Genetic analysis of male-fertility restoration in wheat: VII. A fertility-inhibiting gene. Crop Sci 32:1414–1420
Du H, Maan SS, Hammond JJ (1991) Genetic analysis of male-fertility restoration in wheat: III. Effects of aneuploidy. Crop Sci 31:319–322
Gerlach WL, Bedbrook JR (1979) Cloning and characterization of ribosomal RNA genes from wheat and barley. Nucleic Acids Res 7:1869–1885
Hagras AA, Kishii M, Tanaka H, Sato K, Tsujimoto H (2005) Genomic differentiation of Hordeum chilense from H. vulgare as revealed by repetitive and EST sequences. Genes Genet Syst 80:147–159. doi:10.1266/ggs.80.147
Hattori N, Kitagawa K, Takumi S, Nakamura C (2002) Mitochondrial DNA heteroplasmy in wheat, Aegilops and their nucleus-cytoplasm hybrids. Genetics 160:1619–1630
Ikeguchi S, Hasegawa A, Murai T, Tsunewaki T (1999) Basic studies on hybrid wheat breeding using the 1BL-1RS translocation chromosome/Aegilops kotschyi cytoplasm system 1. Development of male sterile and maintainer lines with discovery of a new fertility-restorer. Euphytica 109:33–42. doi:10.1023/A:1003689100815
Lukaszewski AJ (1994) Manipulation of the genome by chromosome breakage. In: Gill BS, Raupp WJ (eds) Proceedings, US-Japan symposium, classical and molecular cytogenetic analysis, March 1994, Manhattan, Kansas, USA, pp 136–139, 21–23
Lukaszewski AJ (1997) Further manipulation by centric misdivision of the 1RS.1BL translocation in wheat. Euphytica 94:257–261. doi:10.1023/A:1002916323085
Lukaszewski AJ, Curtis CA (1993) Physical distribution of recombination in B-genome chromosomes of tetraploid wheat. Theor Appl Genet 86(1):121–127. doi:10.1007/BF00223816
Maan SS (1992) Genetic analysis of male-sterility restoration in wheat: IV. Fertile line without major Rf genes. Crop Sci 32:24–28
Maan SS, Lucken K (1967) Additional cytoplasmic male sterility-fertility restoration systems in Triticum. Wheat Inf Serv 23:6–9
Maan SS, Lucken KA, Bravo JM (1984) Genetic analyses of male-fertility restoration in wheat I. Chromosome location of Rf genes. Crop Sci 24:17–20
Martín A, Martínez C, Rubiales D, Ballesteros J (1996) Tritordeum: triticale’s new brother cereal. In: Güedes-Pinto H, Darvey N, Carnide VP (eds) Triticale: today and tomorrow. Kluwer, Dordrecht, pp 57–72
Martín A, Martín LM, Cabrera A, Ramírez MC, Giménez MJ, Rubiales D, Hernández P, Ballesteros J (1998) The potential of Hordeum chilense in breeding Triticeae species. In: Jaradat AA (ed) Triticeae III. Science, Enfield, pp 377–386
Martín AC, Atienza S, Ramírez M, Barro F, Martín A (2008a) Male fertility restoration of wheat in Hordeum chilense cytoplasm is associated with 6HchS chromosome addition. Aust J Agric Res 59:206–213. doi:10.1071/AR07239
Martín AC, Atienza SG, Barro F (2008b) Use of ccSSR markers for the determination of the purity of alloplasmic wheat in different Hordeum cytoplasms. Plant Breed 127:470–475. doi:10.1111/j.1439-0523.2007.01483.x
Mcintosh RA, Yamazaki Y, Devos KM, Dubcovsky J, Rogers WJ, Appels R (2003) Catalogue of gene symbols for wheat. In: Proceedings of 10th international wheat genetics symposium. Paestum, Italy, p 4
Murai K (2001) Factors responsible for levels of male sterility in photoperiod-sensitive cytoplasmic male sterile (PCMS) wheat lines. Euphytica 117:111–116. doi:10.1023/A:1004031304997
Murai K (2002) Comparison of two fertility restoration systems against photoperiod-sensitive cytoplasmic male sterility in wheat. Plant Breed 121:363–365. doi:10.1046/j.1439-0523.2002.720110.x
Nasuda S, Kikkawa Y, Ashida T, Rafiqul AKM, Sato K, Endo TR (2005) Chromosomal assignment and deletion mapping of barley EST markers. Genes Genet Syst 80:357–366. doi:10.1266/ggs.80.357
Padilla JA, Martín A (1983) Morphology and cytology of Hordeum chilense X Hordeum bulbosum hybrids. Theor Appl Genet 65(4):353–355. doi:10.1007/BF00276577
Pestsova EG, Ganal MV, Röder MS (2000) Isolation and mapping of microsatellite markers specific for the D genome of bread wheat. Genome 43:689–697. doi:10.1139/gen-43-4-689
Prieto P, Ramírez MC, Ballesteros J, Cabrera A (2001) Identification of intergenomic translocations involving wheat, Hordeum vulgare and Hordeum chilense chromosomes by FISH. Hereditas 135:171–174. doi:10.1111/j.1601-5223.2001.t01-1-00171.x
Raupp WJ, Friebe B, Gill BS (1995) Suggested guidelines for the nomenclature and abbreviation of the genetic stocks of wheat and its relatives. Wheat Inf Serv 81:50–55
Robertson WRB (1916) Chromosome studies. I. Taxonomic relationships shown in the chromosomes of Tettigidae and Acrididae. V-shaped chromosomes and their significance in Acrididae, Locustidae and Gryllidae: chromosome and variation. J Morphol 27:179–331. doi:10.1002/jmor.1050270202
Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal MV (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Sage GCM (1976) Nucleo-cytoplasmic relationship in wheat. Adv Agron 28:265–298
Sears ER (1952) Misdivision of univalents in common wheat. Chromosoma 4:535–550. doi:10.1007/BF00325789
Thomas HM, Pickering RA (1985) Comparisons of the hybrids Hordeum chilense X Hordeum vulgare, Hordeum chilense X Hordeum bulbosum, Hordeum chilense X Secale cereale and the amphidiploid of Hordeum chilense X Hordeum vulgare. Theor Appl Genet 69(5–6):519–522. doi:10.1007/BF00251097
Tsunewaki K (1980) Basic studies on hybrid wheat breeding utilizing the timopheevi cytoplasm and Rf3 gene—summary of the results. Seiken Ziho 29:40–56
Tsunewaki K (1993) Genome-plasmon interaction in wheat. Jpn J Genet 68:1–34. doi:10.1266/jjg.68.1
Wilson JA (1984) Hybrid wheat breeding and commercial seed development. Plant Breed Rev 2:303–319
Wilson P, Driscoll CJ (1983) Hybrid wheat. In: Frankel R (ed) Heterosis. Monographs on theoretical and applied genetics, vol 6. Springer, Berlin Heidelberg New York, pp 94–123
Zhang LL, Zhang YJ (2001) A comparative study on wheat CMS lines with Aegilops juvenalis and Ae. kotschyi cytoplasm. J Hubei Agric Coll 21:193–195
Zhang AM, Nie XL, Liu DC, Guo XL (2001) Advances of hybrid wheat breeding in China. Cereal Res Commun 29:343–350
Acknowledgments
We thank Dr. P. Lazzeri (Agrasys S.L.) for revision and correction of the English in this manuscript. This work was supported by MICINN (Ministerio de Ciencia e Innovación) projects AGL2006-07703 and AGL2007-65685-C02-01 of the Spanish Government.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Martín, A.C., Atienza, S.G., Ramírez, M.C. et al. Chromosome engineering in wheat to restore male fertility in the msH1 CMS system. Mol Breeding 24, 397–408 (2009). https://doi.org/10.1007/s11032-009-9301-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-009-9301-z