Abstract
Rice bacterial leaf blight (BB) caused by Xanthomonas oryzae pv. oryzae and bacterial leaf streak (BLS) caused by X. oryzae pv. oryzicola (Xoc) are two important diseases of rice that often outbreak simultaneously and constrain rice production in much of Asia and parts of Africa. Developing resistant cultivars has been the most effective approach to control BB, however, most single resistance genes have limited value in breeding programs because of their narrow-spectrum of resistance to the races of the pathogen. By contrast, there is little progress in breeding varieties resistant to Xoc since BLS resistance in rice was a quantitative trait and so far only a few quantitative resistance loci have been identified. We reported here the development of a high yield elite line, Lu-You-Zhan highly resistant to both BB and BLS by pyramiding Xa23 with a wide-spectrum resistance to BB derived from wild rice and a non-host maize resistance gene, Rxo1, using both marker assisted selection (MAS) and genetic engineering. Our study has provided strong evidence that non-host R genes could be a valuable source of resistance in combating those plant diseases where no single R gene controlling high level of resistance exists and demonstrated that MAS combined with transgenic technologies are an effective strategy to achieve high level of resistance against multiple plant diseases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rice bacterial leaf blight (BB) caused by Xanthomonas oryzae pv. oryzae (Xoo) is one of the most widely distributed and devastating rice diseases worldwide (David et al. 2006). Bacterial leaf streak (BLS) caused by X. oryzae pv. oryzicola (Xoc) is another serious disease of emerging importance that constrains rice production in certain rice growing regions in China and South/Southeast Asia (Tang et al. 2000). The epidemic of both BB and BLS of rice requires similar environmental conditions and thus the two diseases often occur and outbreak simultaneously in many rice fields of the Central and South China, resulting frequently in heavy yield losses of rice (Zeng and Lin 2003).
For controlling the diseases, developing resistant cultivars is the most economic and effective method (Ogawa 1993). To date, at least 30 resistance (R) genes to BB have been registered (David et al. 2006). Of these, Xa3 and Xa4 have been widely used in breeding programs worldwide and played an important role in controlling the disease in Asia since 1970 s. Currently, almost all commercial indica hybrid and inbred cultivars in China are known to have Xa4, whereas Xa3 is common in japonica varieties (Zhang 1991). This narrow genetic basis for resistance to BB in Chinese rice cultivars has imposed strong selection on the pathogen population, resulting in a dramatic increase in frequency of pathotype V that is virulent to both Xa4 and Xa3 during the past decade in China (Zeng et al. 2002). A different R gene, Xa21 from a wild rice species, O. longistaminata has a broad resistance-spectrum to BB races (Khush et al. 1990). This R gene was cloned (Song et al. 1995) and thus been widely used in rice breeding programs by conventional breeding approach, marker-assisted selection (MAS) and genetic transformation (Tu et al. 1998; Zhai et al. 2001). Unfortunately, Xa21 was again broken down recently by new virulent races in Philippines, Korea, India and China (Lee et al. 1999; Marella et al. 2001; Zeng et al. 2002). In searching for new R genes conferring stable resistance to BB, Xa23, a single completely dominant R gene identified from a wild rice species of O. rufipogon emerged as the most promising one, as it has a broad spectrum of resistance to all tested Chinese BB races during all developmental stages of rice (Zhang et al. 1998, 2001).
In contrast, no single R genes controlling high level of resistance to BLS have been identified in the primary gene pool of rice (Zhao et al. 2004), even though both BB and BLS are caused by two highly related bacterial species of Xanthomonas oryzae (Vauterin et al. 1995). BLS resistance in rice was reportedly a quantitative trait (Xu et al. 1997; Tang et al. 2000). Although the recent identification of quantitative resistance loci (QRLs) has provided new opportunities for improving BLS resistance through MAS (Tang et al. 2000; Zheng et al. 2005; Chen et al. 2006), no MAS efforts have been taken to transfer any identified QRLs to improve BLS resistance in breeding programs because of the complicated nature of QRLs (Li 2001; Li et al. 2006). Recently, a single maize dominant R gene, Rxo1, is reportedly to confer a high level of non-host resistance to rice BLS caused by Xoc (Zhao et al. 2004, 2005), providing an alternative strategy for breeding resistance to BLS.
Here we report an effort in developing elite rice variety resistant to both BB and BLS using combined approaches of phenotypic selection, MAS of Xa23 from wild rice and Agrobacterium tumefaciens mediated transformation of the maize R gene, Rxo1, for BLS resistance.
Materials and methods
Plant materials
A rice line, CBB23-2 carrying Xa23, was used as the donor for resistance to BB and a commercial indica variety, Lu-You-Zhan (LYZ) was used as the recipient. LYZ is a high yielding cultivar in South China with excellent grain quality, but is highly susceptible to both BB and BLS. CBB23-2 was a BC4F5 progeny derived from a cross between an O. rufipogon accession (RBB16, the Xa23 donor) and a recipient indica line, Jingang 30 which is highly susceptible to virtually all Chinese Xoo races (Zhang et al. 2001).
Maker-assisted transfer of the Xa23 gene into rice variety LYZ
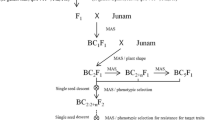
CBB23-2 was used as the male donor and crossed with LYZ to obtain the F1 plants. The true F1 plants were backcrossed to LYZ to obtain the BC1F1 plants. Resistant BC1F1 plants determined by MAS and artificial inoculation were further backcrossed to LYZ to produce the BC2F1 population. Consecutive backcrossing was carried out in the same way to produce BC3F1 and BC4F1 populations. The BC4F1 plants were then selfed twice to produce BC4F3 families. In each BC generation, resistant plants carrying Xa23 were determined by MAS using an SSR marker, RM206 which is 1.9 cM from the Xa23 locus (Pan et al. 2003; http://www.gramene.org), and by artificial inoculation at the 4–5 leaf stage with the most virulent Chinese Xoo strain, GD1358. In the MAS process, the RM206 genotypes were used to infer the genotypes of BC progeny at the Xa23 locus. Finally, one BC4F5 line carrying the homozygous Xa23 gene and LYZ agronomic phenotype was identified and designated as LYZ-Xa23, which was used as the recipient for transformation of Rxo1.
Vector with Rxo1 and rice transformation
The plasmid pCAMBIA1305-1, which contains the whole Rxo1 gene in an approximately 9 kb EcoR I fragment, was kindly provided by Dr. SH Hulbert of Kansas State University, USA. In this study, the binary vector pMNDRBBin6 was used for producing selectable marker-free transgenic plants (Lu et al. 2001), a digested ScaI-Rxo1-NgoMIV fragment of Rxo1 from pCAMBIA1305-1 was ligated into the XmaI-Hpa digested pMNDRBBin6 to obtain the recombinant plasmid pMNDRBBin6-Rxo1, which has the structure of RB1-hph-RB2-Rxo1-LB (Fig. 1) (Xu et al. 2008). pMNDRBBin6-Rxo1 is a double right-border (DRB) vector carrying two copies of T-DNA right-border (RB) sequences flanking the selectable gene, hygromycin phosphotransfase (hph), followed by the Rxo1 gene and one copy of the left border (LB)sequence. Two types of T-DNA inserts, one initiated from the first RB containing the selectable gene and Rxo1 gene, and the other from the second RB containing only Rxo1, were produced and integrated into the genome. The vector pMNDRBBin6-Rxo1 was introduced into an A. tumefaciens strain, EHA105 through electroporation and used in rice transformation. The LYZ-Xa23 plants were used as the recipients of the Rxo1 gene, as described above. Rice transformation was carried out according to the method described by Zhai et al. (2000).
Diagram of the plasmid of binary vector, pMNDRBBin6-Rxo1 Double right-border (DRB) binary vector pMNDRBBin6 described in detail by Lu et al. (2001). RB1, the right border 1 of T-DNA; hph, hygromycin phosphotransfase gene; N, terminator of nos gene; P, CMV35S promoter; RB2, the right border 2 of T-DNA; Rxo1, the Rxo1 gene; LB, the left border of T-DNA. The Sca I and NgoM IV fragment containing 8.5 kb Rxo1 gene was excised from pCAMBIA1305-1 and ligated into the site HpaI and Xma I of pMNDRBBin6 to produce pMNDRBBin6-Rxo1
Molecular analysis of transgenic plants
Two primer pairs were used for molecular analyses of transgenic plants. The first pair from the Rxo1 gene sequence was R1 (5′-ACTATCGGCGAGTACTTCTACACAG-3′) and R2 (5′ - GAGTTTAGCGAGAGCCTGACCTAT-3′) with a 1.45 kb amplified fragment. The second one designed for specific PCR amplification of hph was H1 (5′- TAGGAGGGCGTGGATATGC-3′) and H2 (5′-TACACAGCCATCGGTCCAGA-3′) with a 1.1 kb amplified fragment. All PCR reactions were carried out in 25 μl mixtures containing 50 ng genomic DNA, 10 mmol/l Tris–HCl pH9.0, 50 mmol/l KCl, 1.8 mmol/l MgCl2, 200 μmol/l dNTPs, 50 ng of each primer, and 1 unit of Taq DNA polymerase. PCR reactions were performed in a PTC-100 M Programmable Thermal Controller (MJ Research Inc.) using the following program: predetermination at 94°C for 5 min; then 94°C for 1 min, 56°C for 1 min, and 72°C for 2 min for 35 cycles; followed by 10 min at 72°C. Electrophoresis of the PCR products were carried out on 1.0% agarose gels in 1 × TAE buffer.
In addition, 5–10 μg genomic DNA was extracted from each transgenic line, digested with EcoR I and electrophoresed on a 0.8% agarose gel. Southern hybridization was carried out with ECLTM Direct Nucleic Acid Labeling and Detection System as described by the manufacturer, Amersham. The probes used were the 1.45 kb and 1.1 kb PCR fragments amplified by primer pairs R1/R2 and H1/H2, respectively for confirmation of the Rxo1 gene and hph in the transgenic plants.
Screening of selectable marker-free transgenic plants with Rxo1
The independent transformants of T0 generation were analyzed with primer pairs R1/R2 and H1/H2 and confirmed by artificial inoculation with Xoc strain GD4. T1 seeds were obtained by selfing the highly resistant and positive plants. Segregating plants of T1 generation were further analyzed by PCR and inoculation. Seeds of the plants with Rxo1 but without hph were harvested. The selectable marker-free transgenic plants were identified from the homozygous resistant lines in T2 generation by artificial inoculation and PCR, and confirmed by Southern hybridization with the probes of Rxo1 and hph.
Artificial inoculation and resistance evaluation
The plants were grown in the screenhouse of the Institute of Crop Sciences (ICS), Chinese Academy of Agricultural Sciences, Beijing, China in the summer of 2003–2006. For evaluating BB resistance, 4–5 uppermost leaves of each plant were inoculated with Xoo strains at the seeding or tillering stage by the leaf-clipping method (Kauffman et al. 1973). The lesion length was measured on all inoculated leaves 2 weeks after inoculation. For evaluating BLS resistance, all transgenic plants were inoculated with Xoc strains GD4, FJ1, HB1, HN4 and HN5 collected from the rice fields in Gaungdong, Fujian, Hubei and Hainan provinces using a pin pricked method at the booting stage (Tang et al. 2000). The inoculated plants were scored for their resistances to BB and BLS according to the standard evaluation system of rice (IRRI 1996) 2 weeks after inoculation when lesions became obvious and stable in the susceptible check, LYZ.
To evaluate resistance of the resultant pyramids with Xa23 and Rxo1, seven Chinese Xoo races (Fang et al. 1990), 10 Philippine Xoo races (Li et al. 2006), and five Xoc strains (GD4, FJ1, HB1, HN4 and HN5) were used to inoculate the pyramiding plants in the ICS screenhouse using the same methods described above.
Results
Inheritance of the Xa23 gene in the F1 and BC1F1 plants
Seventeen F1 plants were obtained from the cross of LYZ × CBB23-2. Genotypic data at SSR marker RM206 indicated that all 17 plants had Xa23. The F1 plants were resistant to all seven Chinese Xoo races with an average lesion length of 0.4 ± 0.3 cm, whereas LYZ was susceptible to all tested strains with an average lesion length of 12.5 ± 1.9 cm. The Xa23 donor, CBB 23-2, had an average lesion length of 0.4 ± 0.2 cm. This result indicated that Xa23 is a completely dominant R gene.
Of the 156 BC1F1 plants, 75 plants were inferred to be heterozygous at the Xa23 locus based on their genotypes at RM206 (Fig. 2a). Inoculation with GD1358 at the tillering stage indicated 74 out of 75 plants were resistant with an average lesion length of 0.6 ± 0.2 cm, indicating a genetic distance of 1.3 cM between RM206 and Xa23 and a high accuracy of 98.7% in our MAS for Xa23 using marker RM206.
Segregation of the Xa23 mediated resistance to BB based on the marker genotypes at RM206 which is tightly linked to Xa23 in BC1F1 (a) and BC4F2 (b) populations derived from the cross between LYZ (the recipient) and CBB23-2 (the donor). Letters S and R represent the susceptible and resistant reactions of plants to strain GD1358 of X. oryzae pv. Oryzea, respectively. The markers on the right (a) and left (b) were the 50 bp ladder (NEB Inc.). The arrow at the 130 bp is Xa23-specific DNA fragment. * denotes the homozygous plants with Xa23
Development of LYZ-Xa23 through MAS
Five plants having Xa23 and phenotypically similar to LYZ were selected from 156 BC1F1 plants based on MAS and inoculation confirmation to backcross to LYZ, resulting in a total of 356 BC2F1 progenies. Six resistant plants phenotypically similar to LYZ were selected from the BC2F1 progeny using the same procedure to further backcross with LYZ, producing 410 BC3F1 progeny. Then, seven plants with Xa23 selected from the BC3F1 progeny using the same methods were continuously backcrossed to LYZ, resulting in 475 BC4F1 progeny. Selfed BC4F2 seeds were harvested from five BC4F1 plants that had Xa23 and LYZ phenotype. Genotyping of 450 BC4F2 progeny from the five selected BC4F1 plants with RM206 revealed all three genotypes at RM206 (Fig. 2b), including 26 plants homozygous for the resistant allele and similar to LYZ in phenotype. Resistance of the 26 resultant BC4F3 lines was further confirmed by inoculation. From these lines, one line phenotypically identical to the control (Table 1), LYZ was selected and designated as LYZ-Xa23.
Production of transgenic plants with the Rxo1 gene
The selected LYZ-Xa23 line was transformed with the binary vector, pMNDRBBin6-Rxo1, and 21 T0 transformants were obtained, 11 of which were true transgenic plants based on the presence of the expected 1.45 kb Rxo1 fragment and 1.1 kb hph fragment in these plants when amplified by primer pairs R1/R2 and H1/H2 (Fig. 3). Six of the T0 transformants only produced the 1.1 kb hph fragment. The remaining four transformants were false positives since they produced neither the Rxo1 fragment and nor the hph fragment. The 11 true transgenic plants exhibited a hypersensitivity-like reaction to the Xoc strain, GD4 with restricted necrotic lesions in the pricking areas 2 days after inoculation, whereas the check LYZ showed rapidly spreading water-soaked lesions. However, these transgenic lines differed slightly in their levels of resistance with mean lesion lengths ranging from 0.31 to 0.85 cm 2 weeks after inoculation.
PCR analysis of the transgenic T0 plants. M represents 250 bp molecular weight marker (Takara Inc.); C1 and C2 represent the negative control of LYZ amplified by primer pairs R1/R2 and H1/H2, respectively. C3 and C4 represent the positive control of pMNDRBBin6-Rxo1 amplified by R1/R2 and H1/H2, respectively. Codes 1–30 represent the transgenic lines. The neighboring odd and even codes were the same plants; the bands of odd codes (the 1.45 kb fragment) and even codes (the 1.1 kb fragment) were amplified by R1/R2 and H1/H2, respectively
Generation of selectable market-free transgenic lines
T1 plants derived from seven of the resistant T0 lines were segregating in resistance. Molecular analyses of these plants with H1/H2 identified a total of eight plants which had the 1.45 kb Rxo1 fragment but no hph fragment, suggesting they were selectable marker-free. These included four plants from line L2, three plants from line L7 and only one plant from line L15, from which five plants with the homozygous Rxo1 gene and LYZ –Xa23 phenotype were selected.
Southern analysis confirmed the results from the PCR analyses that there was the DNA band corresponding to the Rxo1 gene (Fig. 4), but no band for hph in the five selected T2 plants. This result indicated that the Rxo1 gene had been stably inherited and the vector backbone sequence was removed in these plants through recombination. The presence of Xa23 in these transgenic lines was further confirmed with RM206. From these, one of the homozygous plants was designated as LYZ-Xa23-Rxo1.
Southern blot analysis of the T2 transgenic plants (lanes 2–6) with the R1/R2 amplified 1.45 kb PCR fragment of Rxo1 from pMNDRBBin6-Rxo1 as the probe. M represents 1 kb molecular weight marker (NEB Inc.); Lanes 1 and 7 represent the digested vector pMNDRBBin6-Rxo1 as the positive control and LYZ as the non-transformed control, respectively
Assessment of the pyramid for resistance to Xoo and Xoc
Table 2 shows the mean lesion lengths of LYZ-Xa23-Rxo1, LYZ-Xa23 and the control LYZ caused by 10 Philippine Xoo races, seven Chinese Xoo races and five Chinese Xoc strains. LYZ was susceptible to all 17 Xoo races and exhibited rapidly spreading water-soaked lesions 24 h after inoculation with an average lesion length reached 8.1 ± 1.7 cm. The LYZ-Xa23-Rxo1 and LYZ-Xa23 lines both showed a high level of resistance to all Xoo races (including the most virulent Chinese race, GD1358) with virtually the same lesion lengths < 1.0 cm, indicating that the presence of Rxo1 didn’t affect the expression of Xa23 in the LYZ-Xa23-Rxo1 line (Fig. 5a). LYZ-Xa23-Rxo1 also showed a high level of resistance to the highly virulent Philippine race 6, PXO99 at the 4–5 leaf stage (data not shown).
Resistant reactions of pyramid line LYZ-Xa23-Rxo1 to Xanthomonas oryzae pv. oryzae (Xoo) and X. oryzae pv. oryzicola (Xoc). a Resistant reactions of LYZ-Xa23-Rxo1 plants to Xoo. Leaves 1 and 2 represent the reactions of susceptible check LYZ and resistant check CBB23-2 to Chinese pathotype V, GD1358; Leaves 3–8 represent the reactions of the LYZ-Xa23-Rxo1 plants to 7 Chinese Xoo pathotypes, JS97-2, KS6-6, JS158-2, Z173, GD1358, OS198 and JS49-6, respectively. b Resistant reactions of LYZ-Xa23-Rxo1 and LYZ (the susceptible check) to Xoc strain GD4 2 weeks after inoculation. Left and right are the leaves of the non-transgenic LYZ and LYZ-Xa23-Rxo1 plant, respectively
When inoculated with the five Xoc strains, LYZ-Xa23-Rxo1 rapidly showed the hypersensitive reaction, and the lesion length of LYZ-Xa23-Rxo1 was 0.27 cm 2 weeks after inoculation (Fig. 5b; Table 2), suggesting that the growth of Xoc in the pyramid was significantly inhibited.
Discussion
To date, most of the 30 reported R genes to BLB have not been used in breeding programs due either to their lower level of resistance or to their narrow spectrum of resistance (David et al. 2006). In this regard, single R genes with wide spectrum of resistance are of choice because of their effectiveness against more Xoo races and the easiness in manipulating target genes in breeding programs. Derived from wild rice O. rufipogen, Xa23 has the broadest spectrum of resistance among all BB R genes identified and confers high level of resistance at all growth stages to the all tested strains, including 10 Philippine races, 7 Chinese pathotypes and 8 isolates from Korea (Zhang et al. 1998, 2001). As demonstrated in this study, the simple inheritance of Xa23 as a single dominant R gene and availability of its closely linked markers make it easy to use Xa23 in breeding programs by either phenotypic selection or MAS. We realize that Xa23 will almost certainly be overcome by new virulent Xoo races sometime in future as long as it becomes widely used in breeding programs according to our knowledge and past experiences (Mew et al. 1992; Adhikari et al. 1995; McDonald and Linde 2002). Fortunately, this can be prevented by combing Xa23 with other R genes such as Xa4, xa5, xa13 and Xa21 to achieve high level and durable resistance to Xoo before it is broken down.
Non-host disease resistance refers to the phenomenon that most plant species are typically resistant to the pathogens of other plant species. This nature of non-host resistance implies that it is of broad spectrum and durable as compared with race-specific R genes of host plants (Heath 1987). As an old phenomenon, non-host resistance has not been used in any breeding programs due to it’s speculated multiple protective mechanisms and the existence of reproductive barriers between different plant species (Thorsten and Volker 2005). Non-host resistance is typically thought to be controlled by multiple genes with general (non-specific) effects on microbial pathogens (Heath 2000). There is, however, growing evidence that single loci can contribute significant effects towards non-host resistance, and some genetic defects that affect active defense processes, like HR, also affect host’s responses to non-host pathogens (Peart et al. 2002; Kang et al. 2003). Thus, there have been increasing interests in non-host resistance and its potential use in heterologous plant species by genetic engineering. The maize Rxo1 gene is able to direct active defense responses that culminate in a rapid HR in a japonica rice (Zhao et al. 2004). Here, we transformed an elite indica rice variety LYZ-Xa23 with Rxo1, further demonstrating that Rxo1 could express stably and produce HR in a heterologous indica background. Our results further indicated that Xa23 and Rxo1 control resistance to BLB and BLS, respectively, and they appeared to function independently.
Achieving a high level of resistance to BLS in the LYZ-Xa23-Rxo1 line in this study was consistent with previously reported results (Zhao et al. 2005) and demonstrated that non-host R genes combined with transgenic technologies are an effective strategy to achieve enhanced resistance against plant diseases in which no high level of resistance controlled by single R genes exists in the host plant species. The strong expression of Rxo1 mediated HR to BLS in both japonica (Zhao et al. 2005) and indica genetic backgrounds demonstrated in this study foresees a wide application of Rxo1 to combat BLS in future rice breeding programs. Although Rxo1 is a single NBS-LRR type of R gene from maize capable of inducing non-host resistance to rice BLS (Zhao et al. 2005), the nature of non-host resistance itself (Heath 1987; Thorsten and Volker 2005) and the lesser degree of differentiation in virulence of the Xoc populations imply that the BLS resistance coffered by Rxo1 might be more durable. However, it would also be very interesting to see how the Xoc populations evolve in responses to Rxo1 after it is widely used in rice breeding programs. Thus, as the major advancement to be achieved in the functional genomic research of major cereals and other model plants (http://www.sanger.ac.uk; http://www.ifti.org; http://www.seqnet.dl.ac.uk) in near future, more and more non-host R genes are expected to be discovered and cloned, providing tremendous opportunities in combating a wide range of plant diseases in agriculture.
Developing superior crop varieties with high level and durable resistance to multiple diseases has been a major challenge to plant breeders and pathologists. In this study, we developed a new high yielding LYZ line pyramided with Xa23 and Rxo1, which represent firstly successful case in achieving this goal using both host and non-host R genes by MAS and genetic engineering. The resultant LYZ-Xa23-Rxo1 line showed a high level of resistance to both BB and BLS, which can be released for commercial use in the epidemic areas of BLB and BLS in Central and South China, or to be used as donor in rice breeding programs for combating BB and BLS within China and elsewhere. Our study has provided additional evidence that non-host R genes could be a valuable source of resistance in combating those plant diseases where no single-R-gene controlled HR exists.
References
Adhikari TB, Vera Cruz CM, Zhang Q, Nelson RJ, Skinner DZ, Mew TW, Leach JE (1995) Genetic diversity of Xanthomonas oryzae pv oryzae in Asia. Appl Environ Microbiol 61:966–971
Chen CH, Zhang W, Huang XM, Lin XH (2006) Major QTL conferring resistance to rice bacterial leaf streak. Agric Sci China 5(3):216–220. doi:10.1016/S1671-2927(06)60041-2
David O, NiÑe L, Pamiela CR, Adam JB (2006) Xanthomonas oryzae pathovars:model pathogens of a model crop. Mol Plant Pathol 7(5):303–324. doi:10.1111/j.1364-3703.2006.00344.x
Fang ZD, Xu ZG, Guo CJ, Yin SZ, Wu SZ, Xu XM, Zhang Q (1990) Studies on pathotypes of Xanthomonas campestris pv. oryzae in China (in Chinese with English abstract). Acta Phytopathol Sin 20(2):81–88
Heath MC (1987) Evolution of plant resistance and susceptibility to fungal invaders. Can J Plant Pathol 9:389–397
Heath MC (2000) Nonhost resistance and nonspecific plant defenses. Curr Opin Plant Biol 3:315–319. doi:10.1016/S1369-5266(00)00087-X
IRRI (1996) Standard evaluation system for rice. IRRI, Manila
Kang L, Li J, Zhao T, Xiao F, Tang X, Thilmony R, He S, Zhou J-M (2003) Interplay of the Arabidopsis nonhost resistance gene NHO1 with bacterial virulence. Proc Natl Acad Sci USA 100:3519–3524. doi:10.1073/pnas.0637377100
Kauffman HE, Reddy APK, Hsieh SPY, Merca SD (1973) A improved technique for evaluation of resistance of rice varieties to Xanthomonas oryzea. Plant Dis Rep 57:537–541
Khush GS, Bacalangco E, Ogawa T (1990) A new gene for resistance to bacterial blight from O.longistamina. Rice Genet Newsl 7:121–122
Lee SW, Choi SH, Han SS, Lee DG, Lee BY (1999) Distribution of Xanthomonas oryzae pv. oryzae strains virulent to Xa21 in Korea. Phytopathology 89:928–933. doi:10.1094/PHYTO.1999.89.10.928
Li ZK (2001) QTL mapping in rice: a few critical considerations. Rice Genetics VI. Proceedings of the fourth international rice genetics symposium, 22–27 Oct., 2000. Science Publishers, Inc./International Rice Research Institute, Los Banos (Philippines), pp 153–171
Li ZK, Arif M, Zhong DB, Fu BY, Xu JL, Domingo-Rey J, Ali J, Vijayakumar CHM, Maghirang R, Yu SB, Khush GS (2006) Complex genetic networks underlying the defensive system of rice (Oryza sativa L.) to Xanthomonas oryzae pv. oryzae. Proc Natl Acad Sci USA 103:7994–7999. doi:10.1073/pnas.0507492103
Lu HL, Zhou XR, Gong ZX, Upadhyaya MN (2001) Generation of selectable marker-free transgenic rice using double right border (DRB) binary vectors. Aust J Plant Physiol 28:241–248
Marella LS, George MLC, Vera CM, Bernardo MA, Nelson RJ, Leung H (2001) Identification of resistance genes effective against rice bacterial blight pathogen in eastern India. Plant Dis 85(5):506–512. doi:10.1094/PDIS.2001.85.5.506
McDonald BA, Linde C (2002) Pathogen population genetics, evolutionary potential, and durable resistance. Annu Rev Phytopathol 40:349–379. doi:10.1146/annurev.phyto.40.120501.101443
Mew TW, Vera Cruz CM, Medalla ES (1992) Changes in the race frequency of Xanthomonas oryzae pv. oryzae in response to rice cultivars planted in the Philippines. Plant Dis 76:1029–1032
Ogawa T (1993) Methods and strategy for monitoring race distribution and identification of resistance genes to bacterial leaf blight (Xanthomonas campetris pv. oryzae) in rice. JARQ 27:71–80
Pan HJ, Wang CL, Zhao KJ, Zhang Q, Fan YL, Zhou SC, Zhu LH (2003) Molecular mapping by PCR-based markers and marker-assisted selection of Xa23 a bacterial blight resistance gene in rice. Acta Agronomica Sin 29(4):501–507
Peart JR, Lu R, Sadanandom A, Malcuit I, Moffet P, Brice DC, Schauser L, Jaggard DA, Xiao S, Coleman MJ, Dow M, Jones JD, Shirasu K, Baulcombe DC (2002) Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc Natl Acad Sci USA 99:10865–10869. doi:10.1073/pnas.152330599
Song WY, Wang GL, Chen LL (1804–1806) Han-Suk Kim, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, Fauquet C, Ronald P (1995) A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science 270:279
Tang DZ, Wu WR, Li WM, Lu H, Worland AJ (2000) Mapping of QTLs conferring resistance to bacterial leaf streak in rice. Theor Appl Genet 101:286–291. doi:10.1007/s001220051481
Thorsten N, Volker L (2005) Non-host resistance in plants: new insights into and old phenomenon. Mol Plant Pathol 6(3):335–345. doi:10.1111/j.1364-3703.2005.00279.x
Tu J, Ona I, Zhang Q, Mew TW, Khush GS, Datta SK (1998) Transgenic rice variety “IR72” with Xa21 is resistant to bacterial blight. Theor Appl Genet 97:31–36. doi:10.1007/s001220050863
Vauterin L, Hoste B, Kersters K, Swings J (1995) Reclassification of Xanthomonas. Int J Syst Bacteriol 45:472–489
Xu J, Wang H, Lin Y, Xi Y (1997) Studies on the inheritance of the resistance of rice to bacterial leaf streak and bacterial leaf blight. Acta Genet Sin 24:330–335
Xu MR, Xia ZH, Zhai WX, Xu JL, Zhou YL, Li ZK (2008) Construction of double right-border binary vector carrying non-host gene Rxol resistant to rice bacterial leaf streak. Chin J Rice Sci 22(2):208–210
Zeng JM, Lin WX (2003) Research progress on rice bacterial leaf streak and its resistance (in Chinese). Mol Plant Breed 1(2):257–263
Zeng LX, Huang SH, Wu SZ (2002) Resistance of IRBB21 (Xa21) to five races of bacterial blight in Guangdong. J Plant Prot 29(2):97–100
Zhai WX, Li XB, Tian WZ, Zhou YL, Pan XB, Cao SY, Zhao XF, Zhao B, Zhang Q, Zhu LH (2000) Introduction of rice blight resistance gene, Xa21, into five Chinese rice varieties through an Agrobacterim-mediated system. Sci China 43(4):361–368 Series C
Zhai WX, Wang WM, Zhou YL, Li XB, Zheng XW, Zhang Q, Wang GL, Zhu LH (2001) Breeding bacterial blight-resistant hybrid rice with the cloned bacterial blight resistance gene Xa21. Mol Breed 8:285–293. doi:10.1023/A:1015234802902
Zhang Q (1991) Genetic evaluation and utilization of resistance to rice bacterial blight in China (in Chinese with English abstract). Scientia Agric Sin 24(2):26–36
Zhang Q, Lin SC, Zhao BY, Wang CL, Yang WC, Zhou YL, Li DY, Chen CB, Zhu LH (1998) Identification and tagging a new gene for resistance to bacterial blight (Xanthomonas oryzae pv. oryzae) from O. rufipogon. Rice Genet Newsl 15:138–142
Zhang Q, Wang CL, Zhao KJ, Zhou YL, Caslana VC, Zhu XD, Li DY, Jiang QX (2001) The effectiveness of advanced rice lines with new resistance gene Xa23 to rice bacterial blight. Rice Genet Newsl 18:71–72
Zhao BY, Ardales E, Brasset E, Claflin LE, Leach JE, Hulbert SH (2004) The Rxo1/Rba1 locus of maize controls resistance reaction to pathogenic and non-host bacteria. Theor Appl Genet 109:71–79. doi:10.1007/s00122-004-1623-y
Zhao BY, Lin XH, Poland J, Trick H, Leach JE, Hulbert SH (2005) A maize resistance gene functions against bacterial streak disease in rice. Proc Natl Acad Sci USA 102(43):15383–15388. doi:10.1073/pnas.0503023102
Zheng JS, Li YZ, Fang XJ (2005) Detection of QTL conferring resistance to bacterial leaf streak in rice chromosome 2 (O. sativa L. spp. indica). Sci Agric Sin 38(9):1923–1925
Acknowledgments
The authors are grateful to the National Natural Science Foundation of China (No. 30571200) and the Ministry of Agriculture (948 # 2006-G51 for providing the financial support. We acknowledge Prof. SH Hulbert, Kansas State University, USA for providing us with plasmid pCAMBIA1305-1 containing Rxo1.
Author information
Authors and Affiliations
Corresponding author
Additional information
Y-L Zhou and J-L Xu contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zhou, YL., Xu, JL., Zhou, SC. et al. Pyramiding Xa23 and Rxo1 for resistance to two bacterial diseases into an elite indica rice variety using molecular approaches. Mol Breeding 23, 279–287 (2009). https://doi.org/10.1007/s11032-008-9232-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-008-9232-0