Abstract
The 3′-UTR of the FAD2 gene from Brassica carinata was cloned by PCR and used to prepare an intron-spliced hairpin RNA (ihpRNA) construct. Compared to that of the wild type (WT) background, this construct, when expressed in B. carinata, resulted in a high degree of FAD2 gene silencing accompanied by strong increases of up to 16 and 10% in oleic acid and erucic acid proportions, respectively. The increase in 18:1 was accompanied by a concomitant proportional reduction in 18:2. A second construct containing ihpRNA targeted to the endogenous FAD2 gene in addition to the heterologous Crambe abyssinica FAE gene under the control of seed specific napin promoter, was used to transform B. carinata. This approach resulted in an even greater increase in erucic acid proportions, by up to 16% in T1 segregating seeds as compared to that of the WT control. To our knowledge, this is currently the highest accumulation of erucic acid achieved in B. carinata seeds using transgenic approaches, making it an increasingly-attractive alternative to high erucic B. napus cultivars as an industrial oil crop.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The strategic goal of our research is to modify high erucic acid (HEA) germplasm of the Brassicaceae to increase the content of erucic acid (22:1Δ13) in the seed oil for industrial niche markets (Jadhav et al. 2005). HEA cultivars are of high interest for industrial purposes because 22:1 is a valuable feedstock with more than 1,000 potential or patented industrial applications (Scarth and Tang 2006). Currently the major derivative of erucic acid is erucamide, which is used as a surface-active additive in coatings and in the production of plastic films as an anti-block or slip-promoting agent. Many other applications are foreseen for erucic acid and its hydrogenated derivative behenic acid, e.g. in lubricants, detergents, film processing agents and coatings, as well as in cosmetics and pharmaceuticals (Leonard 1993; Derksen et al. 1995; Basra and Randhawa 2002; McVetty and Scarth 2002; Puyaubert et al. 2005; Mietkiewska et al. 2007). For many of these industrial uses, the economics are limited by the proportion of 22:1 in the seed oil. With respect to the market for high erucic acid, it is estimated that about 80,000 tons of high erucic oil is used annually worldwide for lubricants, plastics, lacquers, and detergents (http://www.gov.mb.ca/agriculture/research/ardi/projects/98-022.html) Additionally, the European market for high erucic acid oil in 2005 was estimated at 55,000 MT with an annual growth rate of 4–5% (http://www.ienica.net/crops/crambe.pdf). A Brassica cultivar containing erucic acid at levels approaching 80% would significantly reduce the cost of producing erucic acid and its derivatives and could meet the forecasted demand for erucic acid as a renewable, environmentally friendly industrial feedstock (Leonard 1994; Taylor et al. 2002; Jadhav et al. 2005).
As we have shown earlier, over-expression of the Crambe abyssinica FAE gene in Brassica carinata resulted in a substantial increase in the proportion of erucic acid in seeds compared to the wild type control (Mietkiewska et al. 2007) (see schematic in Fig. 1). The synthesis of erucic acid in transgenic B. carinata plants was probably, in part, limited by the smaller microsomal pool of oleoyl-moieties (7–8%) available for elongation. As pointed out previously by Bao et al. (1998) and subsequently by Jadhav et al. (2005) the flux of 18:1 through distinct intermediate lipid pools before elongation might be a factor that limits the availability of 18:1 for elongation. The oleate desaturase, FAD2, is one of the crucial enzymes for the production of polyunsaturated fatty acids in plants (Okuley et al. 1994). As we have shown, by altering the level of FAD2 gene expression using antisense and cosuppression approaches, it was possible to increase the pool of 18:1 available for elongation to enhance production of erucic acid in B. carinata seeds (Jadhav et al. 2005). However, the antisense and cosuppression strategies have variable and unpredictable effectiveness and require the production of large populations of transgenic plants to obtain a reasonable number of lines showing sufficient levels of target gene suppression (Liu et al. 2002).
Simplified schematic of fatty acyl metabolism and triacylglycerol (TAG) assembly in oilseeds, showing the salient ER-based reactions affected by the transformations performed in the present study. The enzymatic steps affected by the Crambe FAE and B. carinata RNAi FAD2 are boxed. The affected pools of acyl-CoAs are circled. The reduction in flux from 18:1 to 18:2 due to the partial silencing of FAD2 which desaturates oleoyl moieties while on PC, is marked by an “X”. The elongation of 18:1 to 20:1 and then 22:1 occurs via fatty acid elongase(s) (FAEs)
The discovery that RNA interference in plants is mediated by sequence-specific degradation of dsRNA has led to the development of highly efficient methods of post transcriptional gene silencing (PTGS). Constructs specially designed to express dsRNA in plants in the form of self-complementary hairpin RNA (hpRNA) elicit a high degree and frequency of PTGS of endogenous genes (http://www.pi.csiro.au/RNAi/, Wesley et al. 2001; Stoutjesdijk et al. 2002). Such hpRNA constructs have great potential for genetic manipulation to improve crop traits (Wang et al. 2000).
We are advocating that B. carinata be developed as an alternative crop platform for industrial oil production and high-erucic oils in particular. B. carinata is easily transformed at very high efficiency (Babic et al. 1998), is highly disease (e.g. blackleg)-resistant, and is drought-resistant, amenable to growth in hotter, drier regions such as the brown soil areas of southern Saskatchewan. While B. carinata has outcrossing rate of 20–30% within its own species, a rate typical Brassicaceae (Murphy 2005), it fortunately also has a very low frequency of out-crossing to canola (Warwick and Black 1993), and therefore poses a lower risk of contaminating oils destined for the food chain. New breeding lines of B. carinata with higher oil and low glucosinolate content are currently being developed at Agriculture and Agri-Food Canada (Dr K. Falk, personal communication) and will provide excellent germplasm for production of high erucic and other industrial oils.
In the present study, we used a partial 3′-UTR of the seed-specific B. carinata FAD2 gene to prepare an intron-spliced hpRNA construct to silence the seed FAD2 gene and consequently, to increase the pool of oleic acid available for elongation. We also demonstrate how an increased pool of oleic acid can contribute to a dramatic increase in the content of erucic acid in Brassica seeds, particularly when combined with heterologous C. abyssinica FAE expression.
Materials and methods
Plant materials and growth conditions
Brassica carinata plants were grown under sterile conditions on MS medium (Murashige and Skoog 1962) during transformation and tissue culture. Transgenic B. carinata plants were grown in the greenhouse at the Kristjanson Biotechnology Complex greenhouses, Saskatoon, SK, under natural light conditions supplemented with high-pressure sodium lamps with a 16 h photoperiod (16 h of light and 8 h of darkness) at 22°C and a relative humidity of 25–30%.
Cloning of 3′-UTR of FAD2 gene
The ORF sequence of seed-specific FAD2-gene (AF124360) from B. carinata was used to design the forward primer 5′-GTCTGCTACGGTCTCTACCG-3′. 3′-RACE was performed using the SMART™ RACE kit (CLONTECH). A cDNA prepared from B. carinata developing seeds was used as a template for PCR amplification during 30 cycles of the following program: 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min. A 686-bp PCR product was cloned into the pCR2.1-TOPO (Invitrogen) cloning vector and subsequently sequenced. Sequence comparison of the PCR product with the FAD2 ORF sequence showed the presence of 236-bp 3′-UTR. The 3′-UTR sequence was submitted to GenBank (accession no: DQ250814).
Gene-silencing constructs
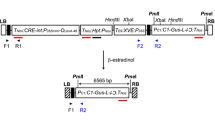
A 142 bp region of the FAD2 3′-UTR was amplified by PCR with primers: 5′-ctcgag GGATGATGATGGTTTAAGA-3′ (lower case shows restriction site for XhoI) and 5′-ggtaccCCATATCACATAATTTAAAGCC-3′ (lower case shows restriction site for KpnI) and cloned in the sense orientation into XhoI and KpnI sites of pKannibal resulting in pKannibal/A plasmid. Subsequently the 3′ UTR was amplified with primers 5′-tctagaGGATGATGATGGTTAAGA-3′ (lower case shows restriction site for XhoI) and 5′-aagcttCCATATCACATAATTTAAAGCC-3′ (lower case shows restriction site for HindIII) and then cloned in the antisense orientation in HindIII and XbaI sites of pKannibal/A giving pKannibal/A-B. The napin promoter (Josefsson et al. 1987) was ligated into pCR2.1 as an XhoI–SacI fragment. Then the XhoI–XbaI cassette carrying intron-interrupted inverted repeats of the FAD2 3′-UTR was excised from pKANNIBAL/A-B and subsequently cloned into the respective sites of pCR2.1 vector (Invitrogen) behind the napin promoter. The resulting plasmid was named XC. A NOS terminator (Bevan 1983) amplified by PCR with primers 5′-tctagaGATCGTTCAAACATTTGGCAA-3′ (lower case shows restriction site for XbaI) and 5′-ggtcgacCGATCTAGTAACATAGATGAC-3′ (lower case shows restriction site for SalI) and subsequently as XbaI–SalI fragment, was ligated with the XbaI–SacI fragment from the XC plasmid into the respective sites of pRD400 (CLONTECH). The resulting plasmid was named XD (Fig. 2).
Schematic diagram (not to scale) of the XD and XS constructs used to transform Brassica carinata plants. Both constructs, driven by napin promoter (Napin P), consist of an inverted repeat of a 142 bp fragment (3′-UTR) corresponding to the 3′-UTR of the B. carinata FAD2 gene (GenBank accession no: DQ250814) separated by the intron of pdk (Wesley et al. 2001). The XS construct also contains the coding region of the C. abyssinica FAE gene (CaFAE). The neomycin phosphotransferase gene (NPTII) is driven by the NOS promoter (NosP). The T-DNA left border (LB) and right border (RB) are shown. The positions of the restriction enzyme sites used for the cloning are as indicated
Isolation of the Crambe abyssinica FAE gene was performed as described previously (Mietkiewska et al. 2007). A C. abyssinica ORF was amplified by PCR with the primers: 5′-cccgggATGACGTTCCATTAACGTAAAG-3′ (lower case restriction site for SmaI) and 5′-ggatccTTAGGACCGACCGTTTTGG-3′ (lower case restriction site for BamHI). The napin promoter was amplified with primers 5′-gaattcAAGCTTTCTTCATCGGTG-3′ (lower case restriction site for EcoRI) and 5′-cccgggGTCCGTGTATGTTTTTAATC-3′ (lower case restriction site for SmaI). The NOS terminator was generated by PCR with the primers 5′-ggatccGATCGTTCAAACATTTGGCAA-3′ (lower case restriction site for BamHI) and 5′-gagctcCGATCTAGTAACATAGATGAC-3′ (lower case restriction site for SacI). The napin promoter as an EcoRI–SmaI fragment, the C. abyssinica FAE as an SmaI–BamHI fragment and the Nos terminator as a BamHI–SacI fragment were ligated into the EcoRI–SacI sites of pSK+, resulting in plasmid ZB. Subsequently the EcoRI–SacI cassette was excited from the ZB plasmid and cloned into the respective sites of the XD plasmid resulting in plasmid XS (Fig. 2).
The final binary vectors XD and XS were electroporated into Agrobacterium tumefaciens cells strain GV3101 containing helper plasmid pMP90 (Koncz and Schell 1986). Plasmid integrity was verified by DNA sequencing following its re-isolation from A. tumefaciens and transformation into E. coli.
Plant transformation
Brassica carinata plants were transformed by the method of Babic et al. (1998). Transgenic plants were selected and analyzed as described by Mietkiewska et al. (2007).
Northern and southern analysis
Total RNA from B. carinata plant material was isolated as described by Lindstrom and Vodkin (1991). Twenty micrograms of RNA was fractionated on a 1.4% (w/v) formaldehyde-agarose gel and the gels were then stained with ethidium bromide to ensure that all lanes had been loaded equally (Sambrook et al. 1989). The RNA was subsequently transferred to Hybond N+ membrane (Amersham Biosciences, Baie d’Urfe, Canada). A 0.5-kb probe containing the 3′ part of FAD2 gene was generated by PCR using primers 5′-GTCTGCTACGGTCTCTACCG-3′ and 5′-TCATAACTTATTGTTGTACCAG-3′ and subsequently radioactively labeled with 32P using a Random Primers Labeling kit (Invitrogen). Membranes were hybridized at 60°C overnight. The filters were washed once in 1× SSPE, 0.1% SDS for 15 min and in 0.1× SSPE, 0.1% SDS for 5–10 min at the temperature of hybridization. The blots were exposed to X-OMAT-AR film (Kodak, Rochester, NY, USA).
Twenty micrograms of B. carinata genomic DNA was digested with the restriction enzyme EcoRI, and the resulting fragments were separated on a 0.9% (w/v) agarose gel and transferred to Hybond N+ nylon membrane via an alkali blotting protocol. For plants transformed with the XD construct, a 1.1-kb DNA fragment containing the napin promoter amplified by PCR using primers 5′-AAGCTTTCTTCATCGGTG-3′ and 5′-TCCGTGTATGTTTTTAATC-3′ was used as the probe. A 1.5-kb probe containing the coding sequence of C. abyssinica FAE was generated by PCR using primers 5′-ATGACGTCCATTAACGTAAAG-3′ and 5′-GGACCGACCGTTTTGGGC-3′ and was used for the analysis of plants transformed with the XS construct. The labeling and hydridization were as described above.
Lipid analyses
The total fatty acid content and acyl composition of B. carinata seed oils were determined by gas chromatography of the fatty acid methyl esters (FAMEs) with 17:0 FAME as an internal standard as described (Katavic et al. 2001; Taylor et al. 2002; Mietkiewska et al. 2007). The lipid class separation was carried out according to the method of Christie (1982). Polar and neutral lipids species were separated by TLC on Silica Gel 60 H plates developed 4 cm in diethyl ether, air dried and then developed in hexane:diethyl ether:acetic acid (70:30:1, v/v/v). Subsequently polar lipids were further developed in chloroform:methanol:acetic acid:water (25:10:3:1, v/v/v). TLC regions containing lipid species were scraped and samples saponified with 2 ml of 10% KOH in methanol at 80°C for 2 h. Following isolation of the free fatty acids, FAMEs were produced using 3 N methanolic HCl and extracted and analyzed by GC as described earlier (Taylor et al. 2002). Relative fatty acid compositions were calculated as the percentage that each fatty acid represented of the total fatty acids. An additional indirect method of assessing the cumulative effects of FAD2 activity during seed fatty acid synthesis through an oleic desaturation proportion (ODP) parameter was calculated as described by Stoutjesdijk et al. (2002).
Results and discussion
Fatty acid composition of preliminary transformants
Brassica carinata plants were transformed with two constructs: XD, targeted at the endogenous FAD2 gene utilizing intron-spliced hpRNA-mediated gene silencing, and XS targeted at silencing the FAD2 gene along with heterologous expression of the Crambe abyssinica FAE gene (Fig. 2). Wild type B. carinata contains two copies of both the FAD2 and FAE genes. Eleven T0 plants carrying the XD construct arising from seven independent transgenic lines, were identified that were both resistant to kanamycin and PCR-positive for the presence of the transgene. Thirty-six plants were transformed with the XS construct representing 20 independent lines (data not shown).
The fatty acid composition of T1 seeds from individual plants was determined for all transformants. The best independent transgenic lines are shown in Fig. 3a. Seed specific silencing of the FAD2 gene resulted in a significant reduction of the level of 18:2 from 19.0% in the wild type background to as low as 4.5% in line XD-4D. The strong reduction in the level of 18:2 was directly correlated with a significant increase in the proportion of oleic acid, up to 21.2% in the best transgenic line carrying the XD construct, compared to 5.7% in the wild type background. The increased pool of 18:1 was utilized by the endogenous microsomal elongation complex (FAE) and this led to the increase in the proportion of erucic acid to as high as 50.6% in line XD-5A, an 11% net increase, and a 26.5% proportional increase over wild-type levels of 40.0% 22:1. The high increase in erucic acid achieved in the current study through ihpRNA-mediated silencing of the FAD2 gene is considerably greater than that reported by Jadhav et al. (2005), where transformation with the FAD2 sequence in an antisense orientation resulted in a net increase in erucic acid of 6% in the best line, compared to the WT B. carinata seeds.
In order to study whether the increased pool of oleic acid can contribute to the higher accumulation of erucic acid obtained by heterologous expression of C. abyssinica FAE, we also designed the XS construct (Fig. 2). As expected, the pronounced silencing of the oleoyl desaturase resulted in large reduction in linoleic acid and concomitant increases in the proportions of oleic acid (Fig. 3b). The level of 18:2 in transgenic lines carrying the XS transgene was as low as 4.0% in XS-18A compared with 16.0% in WT. The increased pool of oleic acid in seeds of the XS transgenic plants was subsequently elongated with assistance from the heterologously-expressed C. abyssinica FAE which resulted in a net increase of the proportion of erucic acid in T1 segregating seeds of up to 16%, a 42% relative increase compared to the wild type. Our current approach resulted in a higher accumulation of erucic acid than that formerly reported utilizing C. abyssinica FAE alone, which resulted in a net increase of 10% in the proportion of erucic acid in T1 seed oils (Mietkiewska et al. 2007).
The effect of gene silencing on oleate desaturation levels in seed from preliminary transformants
Oleate desaturase is highly active in developing seeds of wild type B. carinata, with 85% of 18:1 being converted to 18:2 and 18:3, for an oleic acid desaturation proportion (ODP) value of 0.85 (Table 1). Many transgenic T1 plants carrying XD and XS constructs showed a considerable reduction in the ODP value, to as low as 0.39, indicating a profound (55%) down-regulation of oleate desaturation. Most transgenic plants have ODP values from 0.4 to 0.7 meaning that only 40–70% of oleic acid produced in developing seeds carrying the silencing constructs was converted to polyunsaturated 18-carbon fatty acids compared with 85% in nontransformed B. carinata seeds. As shown in Fig. 4 a positive correlation between the degree of FAD2 gene silencing (expressed as ODP value) and the content of erucic acid was observed. In our study, as in others (Lu et al. 2002; Stoutjesdijk et al. 2002), all transgenic plants (100%) analyzed showed some degree of FAD2 gene silencing (Table 1), a major advantage for this technique over other silencing methodologies wherein only some transformants display the expected phenotypic change. While the intron-spliced hpRNA-mediated (iHP) gene silencing technique used in the present study has not been a means to achieve a complete knockout of the seed oleate desaturase (see http://www.pi.csiro.au/RNAi/; Stoutjesdijk et al. 2002; P Waterhouse, personal communication), iHP did modify the expression of the B. carinata FAD2, with a much higher efficacy of gene silencing than we reported earlier using antisense or cosuppression approaches (Jadhav et al. 2005). In A. thaliana the highest efficiency of FAD2 gene silencing was achieved using an iHP construct utilizing a 120-bp fragment of FAD2 3′-UTR (Stoutjesdijk et al. 2002). The presence of the intron in an iHP construct may result in increased or more stable transcript levels than in the nonintron-containing hpRNA constructs (Tanaka et al. 1990; Stoutjesdijk et al. 2002).
The stability and inheritance of ihpRNA-mediated gene silencing for lipid metabolism related genes has been shown (Stoutjesdijk et al. 2002; Liu et al. 2002); specifically, two published papers on the application of an ihpRNA approach for silencing FAD2 (Yang et al. 2006) and DGAT (Zhang et al. 2005) were based on the results from T0 plants. The high efficiency of gene silencing obtained through the use of ihpRNA-mediated PTGS makes this technique a very valuable contribution to practical trait modification in agricultural plants. This is particularly important for plants with low efficiency of transformation (Liu et al. 2002).
Effect of silencing construct on the FAD2 mRNA level
Based on high oleic acid levels, the six best lines were selected to examine the FAD2 mRNA level in transgenic seeds by northern analysis (Fig. 5). In all transgenic lines, the FAD2 mRNA level decreased compared to that of the wild type (WT). The respective FAD2 3′UTR transgene copy numbers were: XD-7A (1), XD-9A (1), XD-10A (2); XS-18A (1), XS-25A (multiple), XS-28A (1). In the case of XS samples, since the FAD2 3′UTR and Crambe FAE are expressed in tandem between the left and right borders, the copy number refers to both the FAD2 and CrFAE transgene copy numbers. The probe used for the XS Southerns was a Crambe FAE.
Northern blot analysis of FAD2 gene expression in nontransformed B. carinata (WT) and in T1 transgenic seeds carrying the XD and XS constructs. Total RNA was isolated from mid developing seeds, blotted and probed with a 32P-labeled 0.5-kb fragment of the FAD2 gene. In the top panel (a), the bars indicate the relative hybridization signal strength (transcript intensity) with the labeled FAD2 probe; signal intensities are integrated and background subtracted with Image J Image Processing and Analysis Software (http://rsb.info.nih.gov/ij/index.html). Signal strengths are reported relative to that in WT which is set at 1.0. The middle panel (b) is the actual northern gel hybridization result. The lower panel (c) indicates ethidium bromide staining of the ribosomal RNA bands, used to estimate equal loading of the lanes on each gel
While there was not an absolute correlation between transgene gene copy number and FAD2 transcript intensity, the two-copy XD-10A and the multiple copy XS-25A, did exhibit the stronger transcript suppression within their respective datasets. The results demonstrated a good correlation between the degree of transcript reduction induced by PTG silencing, and the increased proportions of the 18:1 in total seed lipids (Fig. 6). Similar trends have been reported from silencing FAD2 gene in tobacco and FAD2-1 in cotton (Yang et al. 2006; Liu et al. 2002).
Effect of silencing constructs on TAG composition in WT vs. XS-18A transgenic B. carinata seeds
Individual lipids extracted from seed tissue were analyzed for fatty acid composition in seeds of WT and of the best performing XS line 18A. In the seeds of XS-18A, both polar and neutral lipids showed an increase in the level of 18:1 compared with WT seeds. Among polar lipids (data not shown), the most pronounced increase of 18:1 was found in phosphatidylethanolamine (PE) and phosphatidylcholine (PC), as high as 6-fold and 3-fold, respectively. In PC from FAD2-silenced seeds, the increase in the level of 18:1 was accompanied by strong reduction in the level of palmitic acid (16:0). Similar changes in polar lipid class composition were reported for leaves and seeds of FAD2 silenced tobacco plants (Yang et al. 2006). These phenomena were also found in an Arabidopsis thaliana FAD2 mutant and in FAD2-silenced Gossypium hirsutum plants (Miquel and Browse 1992; Liu et al. 2002). As proposed by Yang et al. (2006) FAD2-silencing may have effect on de novo 16:0 synthesis. The profound increase of 18:1 with the concomitant decrease of 18:2 may affect the fluidity of membranes, which could result in down-regulation of 16:0 content by a mechanism which is not yet understood.
More importantly, as shown in Table 2, the 18:1 proportion in seed triacylglycerols (TAGs) increased by 3.5-fold compared to the WT seeds, at the expense of 18-carbon polyunsaturated fatty acids. The erucic acid proportion rose from 40% in WT, to 56% in the RNAi FAD2 + Crambe FAE transgenic line XS-18A. This is the best result observed in transgenic manipulations of B carinata to enhance erucic acid content of its seed oil. Such prototypic results make B. carinata an increasingly-attractive alternative to HEA B. napus cultivars as an industrial oil crop.
References
Babic V, Datla RS, Scoles GJ, Keller WA (1998) Development of an efficient Agrobacterium-mediated transformation system for Brassica carinata. Plant Cell Rep 17:183–188. doi:10.1007/s002990050375
Bao X, Pollard M, Ohlrogge J (1998) The biosynthesis of erucic acid in developing embryos of Brassica rapa. Plant Physiol 118:183–190. doi:10.1104/pp.118.1.183
Basra AS, Randhawa LS (2002) Quality improvement in field crops. The Haworth Press, Inc., New York
Bevan M (1983) Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res 12:8711–8721. doi:10.1093/nar/12.22.8711
Christie WW (1982) Lipid analysis, 2nd edn. Pergamon Press, Oxford, England
Derksen JTP, Cuperus FP, Kolster P (1995) Paints and coatings from renewable resources. Ind Crops Prod 3:225–236. doi:10.1016/0926-6690(94)00039-2
Jadhav A, Katavic V, Marillia E-F, Giblin EM, Barton DL, Kumar A et al (2005) Increased levels of erucic acid in Brassica carinata by co-suppression and antisense repression of the endogenous FAD2 gene. Metab Eng 7:215–220. doi:10.1016/j.ymben.2005.02.003
Josefsson LG, Lenman M, Ericson ML, Rask L (1987) Structure of a gene encoding the 1.7 S storage protein, napin, from Brassica napus. J Biol Chem 262:12196–12201
Koncz C, Schell J (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes by a novel type of Agrobacterium binary vector. Mol Gen Genet 204:383–396. doi:10.1007/BF00331014
Leonard EC (1993) High-erucic vegetable oils. Ind Crops Prod 1:119–123. doi:10.1016/0926-6690(92)90009-K
Leonard C (1994) Sources and commercial applications of high erucic vegetable oils. Lipid Tech 4:79–83
Lindstrom JT, Vodkin LO (1991) A soybean cell wall protein is affected by seed color genotype. Plant Cell 3:561–571
Liu Q, Singh SP, Green AG (2002) High-stearic and high-oleic Cottonseed oils produced by hairpin RNA-mediated post transcriptional gene silencing. Plant Physiol 129:1732–1743. doi:10.1104/pp.001933
McVetty PBE, Scarth S (2002) Breeding for improved oil quality in Brassica oilseed species. J Crop Prod 5:345–369. doi:10.1300/J144v05n01_14
Mietkiewska E, Brost JM, Giblin EM, Barton DL, Taylor DC (2007) Cloning and functional characterization of the Fatty Acid Elongase 1 (FAE1) gene from high erucic Crambe abyssinica cv. Prophet. Plant Biotechnol J 5:636–645. doi:10.1111/j.1467-7652.2007.00268.x
Miquel MF, Browse JA (1992) Arabidopsis mutants deficient in polyunsaturated fatty acid synthesis. J Biol Chem 267:1502–1509
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:493–497. doi:10.1111/j.1399-3054.1962.tb08052.x
Murphy DJ (ed) (2005) Plant lipids: biology, utilization and manipulation. Blackwell Publishing, Oxford, UK
Okuley J, Lightner J, Feldmann K, Yadav N, Lark E, Browse J (1994) Arabidopsis FAD2 gene encodes the enzymes that is essential for polyunsaturated lipid synthesis. Plant Cell 6:147–158
Puyaubert J, Garcia C, Chevalier S, Lessire R (2005) Acyl-CoA elongase, a key enzyme in the development of high-erucic acid rapeseed? Eur J Lipid Sci Technol 107:263–267. doi:10.1002/ejlt.200590024
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Scarth R, Tang J (2006) Modification of Brassica oil using conventional and transgenic approaches. Crop Sci 46:1225–1236. doi:10.2135/cropsci2005.08-0245
Stoutjesdijk PA, Singh SP, Liu Q, Hurlstone CJ, Waterhouse PA, Green AG (2002) hpRNA-mediated targeting of the Arabidopsis FAD2 gene gives highly efficient and stable silencing. Plant Physiol 129:1723–1731. doi:10.1104/pp.006353
Tanaka A, Mita A, Ohta S, Kyozuka J, Shimamoto K, Nakamura K (1990) Enhancement of foreign gene expression by a dicot intron in rice but not in tobacco is correlated with an increased level of mRNA and efficient splicing of the intron. Nucleic Acids Res 18:6767–6770. doi:10.1093/nar/18.23.6767
Taylor DC, Katavic V, Zou J, Mackenzie SL, Keller WA, An J, Friesen W, Barton DL, Pedersen KK, Giblin EM, Ge Y, Dauk M, Sonntag C, Luciw T, Males D (2002) Field testing of transgenic rapeseed cv. Hero transformed with a yeast sn-2 acyltransferase results in increased oil content, erucic acid content and seed yield. Mol Breed 8:317–322. doi:10.1023/A:1015234401080
Wang MB, Abbott D, Waterhouse PM (2000) A single copy of a virus derived transgene encoding hairpin RNA gives immunity to barley yellow dwarf virus. Mol Plant Pathol 1:401–410. doi:10.1046/j.1364-3703.2000.00038.x
Warwick SI, Black LD (1993) Guide to the wild germplasm of Brassica and allied crops, Part III: interspecific and intergeneric hybridization in the tribe Brassiceae (Cruciferae). Technical bulletin 1993-16E. Center for Land and Biological Resources Research, Agriculture and Agri-Food Canada
Wesley SV, Helliwell CA, Smith NA, Wang M, Rouse DT, Liu Q et al (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27:581–590. doi:10.1046/j.1365-313X.2001.01105.x
Yang M, Zheng G, Zhang F, Xu Y (2006) FAD2-silencing has pleiotropic effect on polar lipids of leaves and varied effect in different organs of transgenic tobacco. Plant Sci 170:170–177
Zhang F-Y, Yang M-F, Xu Y-N (2005) Silencing of DGAT1 in tobacco causes a reduction in seed oil content. Plant Sci 169:689–694. doi:10.1016/j.plantsci.2005.05.019
Acknowledgments
We are grateful to CSIRO Plant Industry in Australia for providing us with the pKANNIBAL vector and Dr P Waterhouse for his advice. The authors thank Drs Suzanne Abrams and Adrian Cutler for their critical reviews of this manuscript. We also thank to D. Schwab, B. Panchuk, I. Roewer and Dr L. Pelcher for primer synthesis and DNA sequencing. This work was supported by a grant No. 20040417 from Saskatchewan Agriculture Development Fund. This is National Research Council of Canada Publication Number 48451.
Author information
Authors and Affiliations
Corresponding author
Additional information
Database: The nucleotide sequence reported in this paper has been submitted to the EMBL/GenBank under accession number DQ250814.
Rights and permissions
About this article
Cite this article
Mietkiewska, E., Hoffman, T.L., Brost, J.M. et al. Hairpin-RNA mediated silencing of endogenous FAD2 gene combined with heterologous expression of Crambe abyssinica FAE gene causes an increase in the level of erucic acid in transgenic Brassica carinata seeds. Mol Breeding 22, 619–627 (2008). https://doi.org/10.1007/s11032-008-9204-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11032-008-9204-4