Abstract
Cancer-associated fibroblasts (CAFs) have been proved to facilitate colorectal cancer (CRC) development, either with boosting chemo-resistance by communicating with CRC cells in the tumor microenvironment. However, the underlying molecular mechanisms remain largely unclear. Relative expressions of FOSL1 and ITGB4, either with their correlations in CRC tissues, were assessed using qRT-PCR analysis. Also, Kaplan–Meier survival analysis was employed for evaluating the prognosis. Identification of CAFs was determined by the detection of specific makers (α-SMA, FAP, and FSP1) using western blot and immunofluorescence staining. Cell proliferation, self-renewal capacity, and cell apoptosis were estimated by CCK-8, sphere-formation, and flow cytometry assays. Transcriptional regulation of FOSL1 on integrin β4 (ITGB4) was confirmed using ChIP and dual-luciferase reporter assays. Increased FOSL1 and ITGB4 in CRC tissues were both positively correlated with the poor prognosis of CRC patients. Interestingly, FOSL1 was enriched in the CAFs isolated from CRC stroma, instead of ITGB4. CRC cells under a co-culture system with CAFs-conditioned medium (CAFs-CM) exhibited increased FOSL1, promotive cell proliferation, and reduced apoptosis, while these effects could be blocked by exosome inhibitor (GW4869). Moreover, CAFs-derived exosomal FOSL1 was validated to enhance proliferative ability and oxaliplatin resistance of CRC cells. Our results uncovered that CAFs-derived exosomes could transfer FOSL1 to CRC cells, thereby promoting CRC cell proliferation, stemness, and oxaliplatin resistance by transcriptionally activating ITGB4.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) has currently become the third most common malignancy and the fourth primary cause of tumor-related death in the world [1]. The mortality rate of CRC patients is declining due to advances in early screening, surgery, radiotherapy, and adjuvant therapy; however, more than 40% of CRC patients still die from tumor metastasis and recurrence [2, 3]. Acquisition of resistance to chemotherapeutic drugs is one of the main causes of CRC treatment failure [2, 4]. Thus, it is necessary to further explore the molecular mechanism of CRC drug resistance to lay a foundation for developing novel therapeutic strategies.
During tumor pathology, the formation of tumor microenvironment (TEM), based on cancer cells, extracellular matrix, and stromal cells, either with other non-cellular components, is well known to be crucial for tumorigenesis [5]. Accordingly, the cellular communications among these different types of cells through several growth factors, hormones, and cytokines, exert a great impact on the proliferation, metastasis, and differentiation of tumor cells [6]. Cancer-associated fibroblasts (CAFs) are one of the most common tumor stromal cells in TEM, showing completely different biological characteristics from normal fibroblasts (NFs) [7]. CAFs not only provide nutritional support for tumor growth, but also promote tumor progression by regulating a series of intracellular responses, such as metabolic and immune responses [7, 8]. In addition, CAFs have also been proved to affect tumor cell behaviors by transferring active factors into tumor cells via exosomes [9]. As an important extracellular vesicle secreted by various cells, exosome is believed to act as a message transmitter for cellular communication, mainly responsible for the delivery of proteins, molecules, lipids, and so on [10]. Nevertheless, the roles and mechanisms of exosomes derived from CAFs in regulating chemo-resistance and stemness of CRC have not been fully understood yet.
Fos-like antigen 1 (FOSL1) is an oncogenic transcription factor that has been reported to play a key role in regulating tumor progression by regulating tumor-related gene expression [11]. The elevation of FOSL1 expression has been observed in various human tumor tissues, such as CRC, gastric cancer, breast cancer, and hepatocellular carcinoma [12,13,14,15]. High expression of FOSL1 is correlated with a poor prognosis in CRC patients [14]. However, why FOSL1 is overexpressed in CRC and how it affects CRC progression and chemo-resistance remain undetermined. Since a recent study suggested that loss of miR-4516 in CAFs promoted breast cancer progression by increasing FOSL1 expression [16], we boldly speculated that FOSL1 might be overexpressed in CAFs and can be transferred into tumor cells through exosomes.
Herein, by detecting the mRNA and protein expression of FOSL1 and integrin β4 (ITGB4) in the collected clinical CRC samples, we showed that FOSL1 and ITGB4 were abnormally elevated in CRC, and their elevation was associated with a poor prognosis of CRC patients. Interestingly, the different distribution of FOSL1 and ITGB4 in CRC tissues was found, and the former was detected both in tumor cells and the stroma, while the latter was mostly found in tumor cells. Accordingly, FOSL1 was significantly enriched in CAFs isolated from CRC tissues, which could also be transferred from CAFs to CRC cells through exosomes, thereby promoting CRC cell proliferation and oxaliplatin resistance. Mechanistically, we showed that FOSL1 promoted the expression of ITGB4 by directly binding to its promoter region. Thus, our findings suggested that exosomal FOSL1 derived from CAFs might be a potential valuable target for CRC therapy.
Materials and methods
Study subjects
Thirty-nine CRC tissues and corresponding noncancerous tissues were gathered from patients diagnosed with CRC without previous radiotherapy or adjuvant chemotherapy during surgery at Zhongshan City People's Hospital (Zhongshan, China) between December 2013 and December 2018. Patients’ informed consents were obtained from all participants prior to enrollment, and all experimental methods conducted in this work were approved by the Ethics Committee of Zhongshan City People's Hospital.
Cell culture
CRC cell lines, HCT116 and SW480, purchased from American Type Culture Collection (ATCC, USA), were culture in DMEM (Gibco, USA) culture medium plus 10% fetal bovine serum (FBS) as well as 100U/mL penicillin/streptomycin (Gibco by Life Technologies, Grand Island, NY, USA), kept at 37 °C in a humidified atmosphere with 5% CO2.
Isolation of NFs and CAFs
Human CAFs and NFs were isolated from CRC and corresponding noncancerous tissues as described previously [17]. The indicated tissues were chopped into small chunks after washing with sterile PBS, then, digested the chunks with collagenase I (0.1%, Sigma-Aldrich, Germany). The digested solution was centrifuged and re-suspended in DMEM (Gibco) to discard tissue debris. After two days of culture in DMEM containing 10% FBS, the suspending cells and tissues were removed. The most of the remaining cells were fibroblasts, epithelial, and macrophages. Epithelial and macrophages would undergo apoptosis after 5 days of culture with the fibroblasts saved. The isolated CAFs and NFs were identified by the detection of specific markers, including α-SMA, FAP, and FSP1, using immunofluorescence.
Plasmid construction and transfection
The short-hairpin RNA (shRNA) targets FOSL1 (sh-FOSL1) was constructed by inserting FOSL1 sequence into a lentivirus vector (pGLVH1, GenePharma, Shanghai, China). The re-constructed lentiviral vector was then transmitted into HEK293T cells, and puromycin (2 μg/mL) was applied to screen cells with stable expression. FOSL1 sequence was inserted into pcDNA3.1 vector (GenePharma, China) to build FOSL1 overexpression plasmid (FOSL1-OE). The silent or overexpression plasmids were transfected into CRC cells using a Lipofectamine 2000 reagent (Thermo Fisher Scientific, USA), according to the manufacturer’s instructions. After 48 h, the transfection efficiency was determined using qRT-PCR.
Quantitative real-time PCR (qRT-PCR) analysis
TRIzol reagent (Invitrogen, USA) was applied to extract RNAs from tissues and cells, and the quality of the isolated RNA was tested before cDNA synthesis using a PrimeScript RT Kit (Takara, Japan). PCR process was conducted on a Real-Time System (ABI 7500, Applied Biosystems, USA) by utilizing a YBR Green kit (Roche, USA). Sequences of the primers used in this study were listed below.
GAPDH: (Forward: 5’-GTCTCCTCTGACTTCAACAGCG-3’ and Reverse: 5’-ACCACCCTGTTGCTGTAGCCAA-3’);
FOSL1: (Forward: 5’-GGAGGAAGGAACTGACCGACTT-3’ and Reverse: 5’-CTCTAGGCGCTCCTTCTGCTTC-3’);
ITGB4: (Forward: 5’-TCTCTCAGAGTGAGCTGGCAG-3’ and Reverse: 5’-TTCAGCAGCTGGTACTCCAC-3’).
Immunofluorescence (IF) analysis
CAFs and NFs were plated on the coverslip and cultured overnight under 37 °C, and then fixed in 4% paraformaldehyde for 30 min. Cells were incubated with 5% BSA for 1 h after three times of washes. Primary antibodies targeting α-SMA (1:200, Cell Signaling Tech., #19245S, USA), FAP (1:500, Abcam, #ab28244, USA) and FSP1 (1:250, Abcam, #ab197896) were used. After incubation with indicated primary antibodies overnight, Alexa Fluor 488 or 546-labeled secondary antibodies (1:200, Abcam) were used to probe primary antibodies for 2 h.
Immunohistochemical analysis
CRC tissues were dewaxed, hydrated, and incubated with Tris–EDTA (10 mM, pH 9.0) for 30 min at 95 °C. Endogenous peroxidase was subsequently blocked by H2O2 (3%) for 15 min. Afterwards, the slices were incubated overnight with primary antibodies against FOSL1 (1:200, sc376148, Santa Cruz, SC, USA) or ITGB4 (1:100, ab168386, Abcam, USA). Next, the slices were incubated with Dako streptabidin-HRP conjugate (1:300, SC) for 30 min, followed by the incubation with DAB peroxidase substrate for 5 min.
Western blot analysis
Cells were lysed using RIPA buffer (Thermo Fisher, USA), and then proteins were isolated. After the determination of protein concentration, 50 µg sample was separated by SDS-PAGE and then transferred to nitrocellulose membranes. The membranes were incubated in a sequential order with low-fat milk (5%, 2 h), primary antibodies (overnight) against α-SMA (1:1000, Cell Signaling Tech., #19245S, USA), FAP (1:2000, Abcam, #ab28244, USA), and FSP1 (1:1000, Abcam, #ab197896), FOSL1 (1:2000, #sc376148, Santa Cruz, SC, USA) or ITGB4 (1:2000, #ab168386, Abcam, USA), and indicated secondary antibodies for 2 h. Signals were detected using an enhanced chemiluminescence reagent (EMD Millipore, USA).
Extraction and identification of exosomes
CAFs and NFs were cultured in six-well plates containing 3 mL serum-free DMEM until the confluence reached to 90%. Then, the exosomes were collected from the culture medium via filtration using a filter (0.22 µm size), followed by ultracentrifugation (100,000 g, 90 min). The concentrated precipitates underwent centrifugation (4 °C, 100,000 g, 60 min).The precipitates are re-suspended in PBS and stored at − 20 °C until use. The morphology of isolated exosomes was detected using a transmission electron microscopy (TEM, JEOL 2100F, Japan).
Dynamic light scattering (DLS)
The distribution of isolated exosomes was determined via DLS by Zetasizer Nano ZS (Malvern Instrument, UK) using the following setting: fixed angle—173°, temperature—25 °C, refractive index—1.450. Data were then analyzed by the program supplied by the manufacturer.
Co-culture of CAFs-derived exosomes and CRC cells
CRC cells were inoculated in six-well plates with DMEM culture medium supplemented with 5% FBS at a density of 5 × 105 cells per well. After cultured overnight at 37 °C, isolated exosomes were added into the culture medium and allowed for another 24 h incubation.
CCK-8 assay
CAFs and NFs were collected after treatment and seeded into 96-well plates containing DMEM (150 µL), making the final concentration of 1 × 105 cells/mL. After overnight of culture at 37 °C, 10 µl CCK-8 reagent (Dojindo, Japan) was added into each well and incubated at 37 °C for another 2 h. Absorbance was measured at 450 nm by a microplate reader (BioRad, USA).
Sphere-formation assay
Treated CRC cells were collected and seeded in non-adherent dishes (Costar, USA) containing serum-free culture medium with epidermal growth factor (10 ng/ mL, Rocky Hill, USA), 0.5% BSA (Sigma-Aldrich), and 2% B27 (Gibco). Cells were cultured in a cell incubator for two weeks, and then cells were photographed using a microscope (Nikon, Japan) and counted manually.
PKH67 staining and exosome internalization
A PKH67 (Green) Fluorescence kit (Sigma, USA) was adopted to label the lipid bilayer. In brief, the extracted exosomes were re-suspended by diluent C (100 µl), followed by the incubation with PKH67 dye solution for 10 min. The labeling was stopped by placing 500 µl serum. The labeled exosomes were then rinsed by PBS for three times and incubated with HCT116 and SW480 cells for 12 h before imaging.
Cell apoptosis analysis
Cells suspension was fixed overnight using 70% pre-cold ethanol at low temperature. Then, cells were washed with PBS, followed by the incubation with propidium/Annexin V-FITC (Sigma-Aldrich) for 20 min. Afterwards, a flow cytometer (Gallios, China) was utilized to evaluate cell apoptosis.
Chromatin immunoprecipitation (ChIP) assay
The enrichment of FOSL1 in ITGB4 promoter area was determined by a ChIP kit (Merck Millipore, USA) following the protocol provided by the manufacturer. Primers were designed based on the sequence of ITGB4 promoter and synthetized by Invitrogen (Shanghai, China).
Luciferase reporter assay
The luciferase reporter plasmid containing TGB4 promoter sequence into the pGL3 vector was obtained from Genechem (Shanghai, China). Cells were seeded in 24-well plates and cultured for 48 h before transfection. The recombinant luciferase reporter plasmid was co-transfected into cells with FOSL1 overexpression plasmid or shFOSPL1 using lipofectamine 2000 reagent (Invitrogen, USA). Luciferase activity was determined using the luciferase reporter assay kit (Promega, USA) 48 h after transfection.
Statistical analysis
Data were organized as mean ± standard deviation (SD) from at least three independent experiments in this study. Graphpad Prism (Version 7.0, USA) was employed in data analysis using a two-tailed Student’s t test or one-way ANOVA. P value less than 0.05 was considered significant.
Results
Correlation among FOSL1 and ITGB4 in CRC tissues
To investigate whether FOSL1 and ITGB4 play a role in CRC tumorigenesis, we firstly detected the mRNA expression levels of FOSL1 and ITGB4 in CRC tissues. As illustrated in Fig. 1A and C, obvious elevated FOSL1 and ITBG4 were observed in CRC tissues, compared to noncancerous tissues. Interestingly, our results were consistent with the public data from The Cancer Genome Atlas (TCGA) database referring to the exact expression of FOSL1 and ITGB4 in CRC samples (Fig. 1F and G). Moreover, the data from Kaplan–Meier survival analysis also validated that CRC patients with relatively high expression of FOSL1 or ITGB4 both exhibited worse prognosis, compare to those with relatively low expression of FOSL1 and ITGB4 (Fig. 1B and D). Subsequently, Spearman’s correlation analysis discovered a significant positive correlation between FOSL1 expression and ITGB4 expression in CRC tissues (Fig. 1E). Hence, these findings suggested that FOSL1 and ITGB4 may both serve as functional roles to be involved in CRC tumorigenesis.
Correlation among FOSL1 and ITGB4 in CRC tissues. (A) Relative FOSL1 mRNA expression was tested by qRT-PCR in 39 CRC tissues and paired noncancerous tissues. (B) Correlation between FOSL1 expression (low expression, n = 18; high expression, n = 21) and five-year survival rate of 39 CRC patients. (C) Relative ITGB4 mRNA expression was tested by qRT-PCR in 39 CRC tissues and paired noncancerous tissues. (D) The five-year survival rate of 39 CRC patients with low (n = 18) or high (n = 21) expression of ITGB4. (E) The correlation between FOSL1 and ITGB4 expression in 39 pairs of CRC tissues was analyzed by Spearman correlation test. Experiments were performed in triplicate, and error bars represent SD of a triplicate set of experiments. (F and G) Analysis of the expression levels of FOSL1 and ITGB4 in CRC samples from TCGA database. Data are shown as mean ± SD. *P, < 0.05, **P, < 0.01, ***P, < 0.001
The enhanced expression of FOSL1 was observed in CAFs isolated from CRC stroma
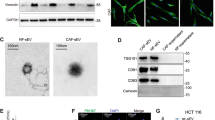
Taking CAFs serving as the crucial elements of surrounding cancer stroma in the microenvironment of tumors into consideration, we subsequently paid our attention to uncover the detailed role of FOSL1 or ITGB4 in CAFs involved in CRC tumorigenesis. First, CAFs from CRC tissues and normal fibroblasts (NFs) from noncancerous tissues were successfully isolated for subsequent experimental preparation. NFs and CAFs elicited fibroblasts characteristics with long fusiform marker morphology, and positive expression patterns of the specific markers, including α-SMA, FAP, and FSP1. As shown in Fig. 2A and B, the expression of α-SMA, FAP, and FSP1 were dramatically increased in CAFs, compared to NFs. In addition, in randomly selected samples of 12 pairs, the expression of FOSL1 mRNA level was also found to be significantly increased in the CAFs compared to NFs (Fig. 2C). Interestingly, subsequent western blot analysis for determining the protein levels of FOSL1 and ITGB4 from random four paired tissues was performed and the data presented that FOSL1 protein level was significantly increased in CAFs compared to NFs, while ITGB4 protein was rarely expressed in NFs, as well as in CAFs (Fig. 2D). These may remind us that FOSL1 could be a main mediator in CAFs.
The enhanced expression of FOSL1 was observed in CAFs isolated from CRC stroma. (A and B) The expression levels of CAFs-specific genes, including α-SMA, FAP, and FSP1, in CAFs and NFs isolated from CRC and paired noncancerous tissues were detected by western blot (A) and immunofluorescence staining (B). (C) Relative FOSL1 mRNA expression in CAFs and NFs was tested by qRT-PCR. (D) Western blot analysis of FOSL1 and ITGB4 in CAFs and NFs. Experiments were performed in triplicate and error bars represent SD of a triplicate set of experiments. Data are shown as mean ± SD. *P, < 0.05, **P, < 0.01, ***P, < 0.001
CAFs-conditional medium accounts for the increased cell proliferation and decreased apoptosis of CRC cells
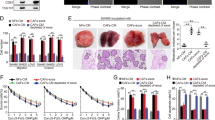
Evidence has shown that CAFs could promote the progression and metastasis of CRC by secreting exosomes to surrounding CRC cells [18]. Thus, we wanted to know whether FOSL1 could be released by CAFs exosome and transferred to CRC cells. We incubated HCT116 and SW480 cells with nothing (blank control), NFs-conditional medium (NFs-CM), or CAFs-conditional medium (CAFs-CM). Compared to blank control and NFs-CM groups, treatment with CAFs-CM strikingly increased the expression of FOSL1 mRNA in HCT116 and SW480 cells (Fig. 3A). Effects of NFs-CM and CAFs-CM treatments on cell viability, self-renewal capacity, and apoptosis were then evaluated by CCK-8, sphere-formation, and flow cytometry assays. As Fig. 3B shown, NFs-CM treatment had no obvious effect on cell viability in comparison with blank control group, while CAFs-CM treatment greatly increased the cell viability compared to NFs-CM group. CAFs-CM treatment was also revealed to increase the number of tumor spheres formed by CRC cells, while NFs-CM treatment had no significant effect on sphere formation (Fig. 3C). Moreover, the apoptosis rate of CRC cells treated with CAFs-CM was significantly decreased compared to NFs-CM treatment, while the apoptosis of CRC cells treated with NFs-CM was not affected (Fig. 3D).
CAFs-conditional medium accounts for the increased cell proliferation and decreased apoptosis of CRC cells. (A) Relative FOSL1 mRNA expression in HCT116 and SW480 cells treated with nothing (blank control), NFs-CM or CAFs-CM. (B) The viability of HCT116 and SW480 cells from each group was evaluated via CCK-8 assay. (C) The self-renewal ability of HCT116 and SW480 cells from each group was assessed by sphere-formation assay. (D) The apoptosis rate of HCT116 and SW480 cells was detected after treatment with nothing, NFs-CM or CAFs-CM. Experiments were performed in triplicate and error bars represent SD of a triplicate set of experiments. Data are shown as mean ± SD. **P, < 0.01, ***P, < 0.001
CAFs-derived exosomes may participate in the enhanced expression of FOSL1, proliferation, and oxaliplatin resistance of CRC cells
To confirm the effects of CAFs-derived exosomes, we treated CRC cells with NFs-CM, CAFs-CM, or CAFs-CM + GW4869 (an inhibitor of exosome). As expected, the increased expression of FOSL1 observed in CAFs-CM treated HCT116 and SW480 cells was abolished by the treatment with GW4869 (Fig. 4A). The promotive effects of CAFs-CM treatment on cell viability of HCT116 and SW480 cells were also reversed by the treatment with GW4869 (Fig. 4B). Moreover, CAFs-CM treatment was found to increase the resistance of HCT116 and SW480 cells to oxaliplatin, while this phenomenon was also blocked by GW4869 treatment (Fig. 4C). By using flow cytometry analysis, we found that oxaliplatin-induced apoptosis of SW480 cells was significantly decreased by CAFs-CM treatment; however, CAFs-CM + GW4869 treatment failed to decrease oxaliplatin-induced apoptosis of SW480 cells (Fig. 4D). Additionally, NFs-CM treatment had no significant effect on ITGB4 expression of HCT116 and SW480 cells, while CAFs-CM treatment strikingly increased the expression of ITGB4 (Fig. 4F). And the CAFs-CM-induced elevation of ITGB4 expression in HCT116 and SW480 cells was reversed by GW4869 treatment (Fig. 4E). These findings suggested that CAFs-derived exosomes might contribute to the increased expression of FOSL1, cell proliferation, and oxaliplatin resistance of CRC cells.
CAF-derived exosomes may participate in the enhanced expression of FOSL1, proliferation, and oxaliplatin resistance of CRC cells. (A) Relative FOSL1 mRNA expression in HCT116 and SW480 cells treated with NFs-CM, CAFs-CM, or CAFs-CM plus GW4869 (exosome inhibitor). (B) The viability of HCT116 and SW480 cells was measured after treatment with NFs-CM, CAFs-CM, or CAFs-CM plus GW4869. (C) The resistance of HCT116 and SW480 cells to oxaliplatin was assessed after treatment with NFs-CM, CAFs-CM, or CAFs-CM plus GW4869 in the presence of oxaliplatin (0, 1, 2, 4, 8, 16 µM). (D) The apoptosis rate of HCT116 and SW480 cells was detected after treatment with NFs-CM, CAFs-CM, or CAFs-CM plus GW4869. (E) Relative ITGB4 mRNA expression in HCT116 and SW480 cells treated with nothing, NFs-CM, CAFs-CM, or CAFs-CM plus GW4869. Experiments were performed in triplicate, and error bars represent SD of a triplicate set of experiments. Data are shown as mean ± SD. *P, < 0.05, **P, < 0.01, ***P, < 0.001
CAFs-derived exosomes transferring FOSL1 to CRC cells thereby boosting cell proliferation and oxaliplatin resistance
To further confirm the role of CAFs-derived exosomes in CRC tumorigenesis, we isolated exosomes from both NFs and CAFs (NFs-exo and CAFs-exo), and the morphology of exosomes was observed. The data from Fig. 5A and B revealed that the morphology of exosomes isolated from NFs and CAFs was spherical with a diameter of about 110 nm. Western blot analysis suggested that the isolated exosomes expressed exosomal surface markers, CD63 and TSG101, and that they poorly expressed negative marker GM130 (Fig. 5C). To confirm the internalization of exosomes into HCT116 and SW480 cells, exosomes labeled with PKH67 green fluorescence were measured under a fluorescence microscope, and the images suggested that green fluorescence was both detected in HCT116 and SW480 cells (Fig. 5D), indicating the internalization of exosomes by CRC cells. The expression of FOSL1 in NFs-exo and CAFs-exo was detected by qRT-PCR. Compared to PBS group, FOSL1 expression was no significant change in NFs-exo, while it was significantly increased in CAFs-exo (Fig. 5E). To investigate whether FOSL1 could be transferred to CRC cells by exosomes, we treated HCT116 and SW480 cells with NFs-exo and CAFs-exo. The results indicated that FOSL1 expression was markedly increased in HCT116 and SW480 cells treated with CAFs-exo, while NFs-exo treatment had no effect on FOSL1 expression (Fig. 5F). In functional assays, we found that CAFs-exo treatment dramatically increased the cell viability (Fig. 5G), the renewal capacity (Fig. 5H), and the resistance to oxaliplatin (Fig. 5I) of CRC cells, while NFs-exo had no significant effect on cell viability, renewal capacity, and resistance to oxaliplatin. In addition, CAFs-exo, but no NFs-exo, treatment was revealed to increase the expression of ITGB4 in HCT116 and SW480 cells (Fig. 5J). These findings suggested that CAFs might contribute to the CRC cell proliferation and oxaliplatin resistance by transferring FOSL1 to CRC cells trough exosomes.
CAFs-derived exosomes transferring FOSL1 to CRC cells thereby boosting cell proliferation and oxaliplatin resistance. (A) TEM observation of exosomes isolated from NFs-CM and CAFs-CM. (B) Particle size analysis of exosomes isolated from NFs-CM (NFs-exo) and CAFs-CM (CAFs-exo). (C) Protein expression of CD63, TSG101, and GM130 in NFs, NFs-exo, CAFs, and CAFs-exo. (D) The internalization of exosomes that labeled with fluorescent PKH67 to HCT116 and SW480 cells was observed by immunofluorescence microscopy. (E) Relative FOSL1 mRNA expression in PBS, NFs-exo, and CAFs-exo. (F) Relative FOSL1 mRNA expression in HCT116 and SW480 cells treated with PBS, NFs-exo, or CAFs-exo. (G) The viability of HCT116 and SW480 cells was tested after treatment with PBS, NFs-exo, or CAFs-exo. (H) Effects of PBS, NFs-exo, or CAFs-exo treatment on self-renewal ability of HCT116 and SW480 cells were estimated by sphere-formation assay. (I) The oxaliplatin resistance of HCT116 and SW480 cells was analyzed using CCK-8 assay after treatment with PBS, NFs-exo, or CAFs-exo. (J) Relative ITGB4 mRNA expression in HCT116 and SW480 cells treated with PBS, NFs-exo, and CAFs-exo. Experiments were performed in triplicate and error bars represent SD of a triplicate set of experiments. Data are shown as mean ± SD. *P, < 0.05, **P, < 0.01, ***P, < 0.001
Correlation among FOSL1 and ITGB4 in CRC tissues
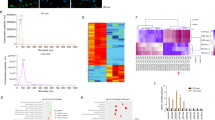
Since FOSL1 could be transferred to CRC cells from CAFs by exosomes, and CAFs-exo treatment could increase the expression of ITGB4 (Fig. 5J), therefore, we speculated that the increased ITGB4 in CRC cells might be caused by exosomal FOSL1. Thus, we designed an overexpression plasmid and a shRNA to overexpress and knockdown FOSL1 expression in HCT116 and SW480 cells. Results from qRT-PCR suggested that, in HCT116 and SW480 cells, FOSL1 overexpression plasmid significantly increased the expression of FOSL1, and sh-FOSL1 transfection successfully decreased the expression of FOSL1 (Fig. 6A). Overexpression of FOSL1 remarkably increases ITGB4 expression and knockdown of FOSL1 markedly decreases ITGB4 expression in HCT116 and SW480 cells (Fig. 6B), indicating that FOSL1 positively regulated the expression of ITGB4. With the prediction of the JASPAR, FOSL1 was revealed to harbor-binding site on the promoter area of ITGB4 (Fig. 6C). To confirm the binding relationship between FOSL1 and ITGB4 promoter, a ChIP assay was conducted. The results revealed an obvious occupancy of FOSL1 at the promoter region of ITGB4 (Fig. 6D). Next, the FOSL1-binding sequence of ITGB4 promoter was constructed into the luciferase reporter plasmids to perform luciferase reporter assay. The data presented that FOSL1 overexpression led to a significant increase of luciferase activity, while knockdown of FOSL1 resulted in a remarkable decrease of luciferase activity (Fig. 6E and F). These results indicated that FOSL1 could promote ITGB4 expression by directly binding to its promoter.
Correlation among FOSL1 and ITGB4 in CRC tissues. (A) The overexpression or knockdown efficiency of FOSL1 overexpression plasmid or sh-FOSL1 was verified by qRT-PCR. (B) Effects of FOSL1 overexpression or knockdown on the expression of ITGB4 in HCT116 and SW480 cells were tested by qRT-PCR. (C) The prediction of FOSL1-binding sites within the promoter area of ITGB4 provided by JASPAR database. (D) ChIP was performed to validate the binding capacity of FOSL1 to the promoter of ITGB4. (E) The interaction between FOSL1 and ITGB4 was tested by luciferase reporter assay in HCT116 and SW480 cells. Experiments were performed in triplicate and error bars represent SD of a triplicate set of experiments. Data are shown as mean ± SD. *P, < 0.05, ***P, < 0.001
Discussion
CAFs have been proved to be functionally heterogeneous with even completely opposite impacts on tumor progression [9]. CAFs display tumor-facilitating functions in most cases, however, in some occasions anti-tumorigenic functions have been reported [9]. Evidence has shown that CAFs play a pivotal role in modulating tumor cell metastasis, drug resistance, invasion, angiogenesis, and immune evasion by communicating with other cells within the TME [19]. Communication between CAFs and tumor cells can be reached through direct cell-to-cell contact or indirect communication. The release of exosomes is a critical indirect communication way of CAFs to affect tumor cell behavior [20, 21]. Abnormal expression of molecules contained in exosomes derived from CAFs can lead to dysregulation of signaling pathways in cancer cells after uptake by cancer cells [22, 23]. Emerging evidences have shown that CAFs are key determinants in regulating growth, metastasis, invasion, and drug resistance of CRC through exosomes-mediated cellular communication [24]; however, the underlying mechanisms remain to be further explored. Most of the studies suggested that non-coding RNAs, such as micro-RNAs (miRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs), are one of the most commonly transferred molecules by exosomes to CRC cells [17, 18, 25, 26]. However, whether other molecules, such as transcription factors, can be transferred from CAFs to CRC cells through exosomes-mediated way remains undetermined.
As an important transcription factor, FOSL1 has been found to be elevated in numerous of human cancers and is associated with lymphatic infiltration, clinical TNM stage, and poor prognosis [27,28,29]. It was also reported to be increased in CRC, and knockdown of FOSL1 could reduce the growth, metastasis, and angiogenesis of CRC cells, suggesting that FOSL1 may be a potential therapeutic target for CRC [14]. However, the expression of FOSL1 in CAFs surrounding CRC cells remain undetermined. Herein, we revealed that FOSL1 was elevated in both CRC tissues and CAFs isolated from CRC tissues. These results indicated that the increased FOSL1 in CRC may be derived from the surrounding CAFs. In the following assays, we demonstrated that exosomes derived from CAFs contributed to the transfer of FOSL1 from CAFs to CRC cells. Unfortunately, we are unable to provide direct evidence that the changes in FOSL1 and ITGB4 in CRC cells after exosome treatment were caused by exosome FOSL1 transport based on the existing data and findings. Because there are many kinds of substances in the exosome, including mRNA, non-coding RNAs, and even proteins. The change of FOSL1 expression in CRC after exosome treatment could also be caused by the transport of specific non-coding RNAs. This could be a direction for our future research, we will try to answer this question in a new project.
ITGB4 has been identified to be a novel promising serum diagnostic biomarker and a potential therapeutic target for CRC [30,31,32]. However, the association between ITGB4 and CAFs in CRC has not been reported. Interestingly, a previous study revealed that ITGB4 was elevated in triple-negative breast cancer cells; moreover, the increased ITGB4 in TNBC cells could be transferred to CAFs by exosomes [33]. Similarly, ITGB4 was found predominantly expressed in hepatocellular carcinoma cells but not in infiltrating stromal cells [34]. In our study, we found that ITGB4 was increased in CRC but not in surrounding CAFs. Interestingly, we found that FOSL1 promoted the expression of ITGB4 by binding to its promoter, suggesting that CAFs contributed to the elevation of ITGB4 indirectly by transferring FOSL1 to CRC cell through exosomes.
In conclusion, our findings suggested that exosomes derived from CAFs transferred FOSL1 to CRC cells, therefore, promoting CRC progression and oxaliplatin resistance by increasing the expression of ITGB4. Our findings suggested that FOSL1 in CAFs may be a therapeutic target for CRC. Nevertheless, there are many shortcomings, such as the precise distribution of FOSL1 and ITGB4 in clinical samples, and whether this regulatory pattern could be also suitable for in vivo studies, which is urgent to be further investigated. Herein, our study was only at the step conducted in vitro and needs to be further verified in vivo, which will be a key in our follow-up study.
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author.
References
Brenner H, Kloor M, Pox CP (2014) Colorectal cancer. Lancet 383(9927):1490–1502
Punt CJ, Tol J (2009) More is less – combining targeted therapies in metastatic colorectal cancer. Nat Rev Clin Oncol 6(12):731–733
Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB (2019) Colorectal cancer. Lancet 394(10207):1467–1480
Van der Jeught K, Xu HC, Li YJ, Lu XB, Ji G (2018) Drug resistance and new therapies in colorectal cancer. World J Gastroenterol 24(34):3834–3848
Arneth, B., Tumor Microenvironment. Medicina (Kaunas), 2019. 56(1).
Xiao Y, Yu D (2021) Tumor microenvironment as a therapeutic target in cancer. Pharmacol Ther 221:107753
Biffi G, Tuveson DA (2021) Diversity and biology of cancer-associated fibroblasts. Physiol Rev 101(1):147–176
Chen Y, McAndrews KM, Kalluri R (2021) Clinical and therapeutic relevance of cancer-associated fibroblasts. Nat Rev Clin Oncol 18(12):792–804
Li C, Teixeira AF, Zhu HJ, Ten Dijke P (2021) Cancer associated-fibroblast-derived exosomes in cancer progression. Mol Cancer 20(1):154
Zhang L, Yu D (2019) Exosomes in cancer development, metastasis, and immunity. Biochim Biophys Acta Rev Cancer 1871(2):455–468
Sobolev VV, Khashukoeva AZ, Evina OE, Geppe NA, Chebysheva SN, Korsunskaya IM et al (2022) Role of the Transcription Factor FOSL1 in Organ Development and Tumorigenesis. Int J Mol Sci 23(3):1521
Zhu X, Liu H, Xu Z, Zhang Y (2019) Expression and clinical significance of FOS-like antigen 1 in gastric adenocarcinoma. Pathol Res Pract 215(6):152394
Duan L, Zhao M, Ang L, Hu H, Wu Z, Wang J et al (2020) Down-regulation of FOS-like antigen 1 enhances drug sensitivity in breast cancer. Int J Clin Exp Pathol 13(8):2092–2099
Liu Y, Yue M, Li Z (2021) FOSL1 promotes tumorigenesis in colorectal carcinoma by mediating the FBXL2/Wnt/beta-catenin axis via Smurf1. Pharmacol Res 165:105405
Li L, Zhang W, Zhao S, Sun M (2019) FOS-like antigen 1 is a prognostic biomarker in hepatocellular carcinoma. Saudi J Gastroenterol 25(6):369–376
Kim JE, Kim BG, Jang Y, Kang S, Lee JH, Cho NH (2020) The stromal loss of miR-4516 promotes the FOSL1-dependent proliferation and malignancy of triple negative breast cancer. Cancer Lett 469:256–265
Zhou L, Li J, Tang Y, Yang M (2021) Exosomal LncRNA LINC00659 transferred from cancer-associated fibroblasts promotes colorectal cancer cell progression via miR-342-3p/ANXA2 axis. J Transl Med 19(1):8
Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen S et al (2018) Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics 8(14):3932–3948
Liao Z, Tan ZW, Zhu P, Tan NS (2019) Cancer-associated fibroblasts in tumor microenvironment - Accomplices in tumor malignancy. Cell Immunol 343:103729
Yang X, Li Y, Zou L, Zhu Z (2019) Role of exosomes in crosstalk between cancer-associated fibroblasts and cancer cells. Front Oncol 9:356
von Ahrens D, Bhagat TD, Nagrath D, Maitra A, Verma A (2017) The role of stromal cancer-associated fibroblasts in pancreatic cancer. J Hematol Oncol 10(1):76
Affo S, Yu LX, Schwabe RF (2017) The Role of Cancer-Associated Fibroblasts and Fibrosis in Liver Cancer. Annu Rev Pathol 12:153–186
Yoshida GJ (2020) Regulation of heterogeneous cancer-associated fibroblasts: the molecular pathology of activated signaling pathways. J Exp Clin Cancer Res 39(1):112
Hu JL, Wang W, Lan XL, Zeng ZC, Liang YS, Yan YR et al (2019) CAFs secreted exosomes promote metastasis and chemotherapy resistance by enhancing cell stemness and epithelial-mesenchymal transition in colorectal cancer. Mol Cancer 18(1):91
Chen X, Liu Y, Zhang Q, Liu B, Cheng Y, Zhang Y et al (2021) Exosomal miR-590-3p derived from cancer-associated fibroblasts confers radioresistance in colorectal cancer. Mol Ther Nucleic Acids 24:113–126
Gu C, Lu H, Qian Z (2020) Matrine reduces the secretion of exosomal circSLC7A6 from cancer-associated fibroblast to inhibit tumorigenesis of colorectal cancer by regulating CXCR5. Biochem Biophys Res Commun 527(3):638–645
Dai C, Rennhack JP, Arnoff TE, Thaker M, Younger ST, Doench JG et al (2021) SMAD4 represses FOSL1 expression and pancreatic cancer metastatic colonization. Cell Rep 36(4):109443
Vallejo A, Erice O, Entrialgo-Cadierno R, Feliu I, Guruceaga E, Perugorria MJ et al (2021) FOSL1 promotes cholangiocarcinoma via transcriptional effectors that could be therapeutically targeted. J Hepatol 75(2):363–376
Zhang M, Hoyle RG, Ma Z, Sun B, Cai W, Cai H et al (2021) FOSL1 promotes metastasis of head and neck squamous cell carcinoma through super-enhancer-driven transcription program. Mol Ther 29(8):2583–2600
Jiang X, Wang J, Wang M, Xuan M, Han S, Li C et al (2021) ITGB4 as a novel serum diagnosis biomarker and potential therapeutic target for colorectal cancer. Cancer Med 10(19):6823–6834
Chen B, Ma Y, Bi J, Wang W, He A, Su G et al (2021) Regulation network of colorectal-cancer-specific enhancers in the progression of colorectal cancer. Int J Mol Sci 22(15):8337
Zhu X, Jiang X, Zhang Q, Huang H, Shi X, Hou D et al (2022) TCN1 Deficiency Inhibits the Malignancy of Colorectal Cancer Cells by Regulating the ITGB4 Pathway. Gut Liver. https://doi.org/10.5009/gnl210494
Sung JS, Kang CW, Kang S, Jang Y, Chae YC, Kim BG et al (2020) ITGB4-mediated metabolic reprogramming of cancer-associated fibroblasts. Oncogene 39(3):664–676
Leng C, Zhang ZG, Chen WX, Luo HP, Song J, Dong W et al (2016) An integrin beta4-EGFR unit promotes hepatocellular carcinoma lung metastases by enhancing anchorage independence through activation of FAK-AKT pathway. Cancer Lett 376(1):188–196
Acknowledgements
The authors thanked the participants for their contribution to the study. We also thanked Prof. Hu for her assistance during data analysis and manuscript revision.
Funding
This work was supported by the Medical Research Foundation of Guangdong Province (No.B2022159); Research Project of Jiangmen Central Hospital (No.J202104) and Medical and Health Science and Technology Project of Jiangmen (No.2022YL01065, No.2022YL01089).
Author information
Authors and Affiliations
Contributions
SSL—acquisition of data; analysis and interpretation of data; drafting the manuscript. BZ—conception and design of study; acquisition of data; revising the manuscript critically for important intellectual content.
Corresponding author
Ethics declarations
Competing interests
The authors have no commercial or other associations that might pose a conflict of interest.
Ethical approval
This study has obtained approval of the Ethics Committee of Zhongshan City People’s Hospital.
Consent to participate
Written informed consent was obtained from the patients.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lin, S., Zhu, B. Exosome-transmitted FOSL1 from cancer-associated fibroblasts drives colorectal cancer stemness and chemo-resistance through transcriptionally activating ITGB4. Mol Cell Biochem 479, 665–677 (2024). https://doi.org/10.1007/s11010-023-04737-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-023-04737-9