Abstract
Increasing evidence indicates that microRNA (miRNA) regulated mechanisms in myocardial healing and ventricular remodeling following acute myocardial infarction (AMI). We aim to comprehensively investigate changes of exosomal miRNA profile during the post-MI period and determine potential miRNAs associated to adverse left ventricular remodeling (ALVR). We prospectively evaluated ST-elevated MI patients with cardiac magnetic resonance imaging at the 2 weeks and 6 months after AMI (n = 10). ALVR was defined as an increase in LV end-diastolic and end-systolic volume > 13%. The blood samples were taken for miRNA measurements at the baseline, 2 and 6 weeks after AMI. In the miRNA profile assessment, 8 miRNAs were identified that were associated ALVR (miR-199a-5p, miR-23b-3p, miR-26b-5p, miR-301a-3p, miR-374a-5p, miR-423-5p, miR-483-5p and miR-652-3p). Three of them (miR-301a-3p, miR-374a-5p and miR-423-5p) differed significantly between patients with and without ALVR during follow-up period and the rest of them during the acute phase of AMI. The detection of these miRNAs, which have different role in various pathways, necessitate future mechanistic studies unravel the complex remodeling process after AMI.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The pathophysiological basis of adverse left ventricular remodeling (ALVR) after acute myocardial infarction (AMI) includes the ionic, genomic, cellular, and extracellular levels changes that occur during the process of wound healing [1]. If cardiac stress conditions are persistent, the process can become irreversible and cellular and extracellular changes such as apoptosis, necrosis, and fibrosis can occur [2]. Depending on these changes, the fate of infarct healing can either be adaptive (reverse) or maladaptive (adverse) remodeling, respectively preserving or worsening of cardiac function and geometry [3]. However, the molecular mechanisms underlying left ventricular remodeling after AMI are not yet fully understood.
MicroRNAs (miRNAs) are non-coding RNA molecules of 19–25 nucleotides that join in gene expression regulation at the post-transcriptional level [4]. Reprogramming of gene expression underlies the pathophysiology of ALVR [5]. Therefore, miRNAs play a key role in physiological-pathological adaptations of heart [6]. miRNAs are expressed in multiple cell types including cardiomyocytes, fibroblasts, endothelial cells and inflammatory cells [7]. It is thought that these miRNAs may have a role in the regulation of basic processes such as excessive myocardial fibrosis, pathological cardiomyocyte hypertrophy and myocardial cell apoptosis. Numerous studies have shown that dysregulation in the expression of microRNA (miRNA) molecules leads to changes in various pathological processes in the heart, which are associated with post-AMI transition from cardiac hypertrophy to heart failure. Previous panel miRNA studies have shown that miR-1, miR-21, miR-24, miR-27a, miR-29 family members (miR-29a and miR-29b), miR-101, miR-133a, and miR-208a/b miRNAs have been implicated in left ventricular dysfunction after AMI [8,9,10,11]. It is thought that these miRNAs may have a role in the regulation of basic processes such as excessive myocardial fibrosis, pathological cardiomyocyte hypertrophy and myocardial cell apoptosis [12].
MiRNAs can be packaged in microparticles such as exosomes. Exosomal miRNAs are more stable in biological fluids compared to plasma miRNAs [13]. It is reasonable to consider that exosomal miRNAs are not released passively from cells selectively loaded into exosomes to serve specific functions, as exosomal miRNAs have a specific expression pattern under altered pathophysiological conditions [14]. However, there are limited studies about the role of exosomal miRNAs in ALVR after AMI. Therefore, an increasing interest emerges in miRNAs as potential diagnostic biomarkers and a new therapeutic target involved in the pathophysiology of AR after AMI. In the present pilot study, we explored the temporal changes of exosomal miRNAs using an extensive assessment protocol in patients after AMI. The aim was to find novel miRNAs of exosomal origin that may have a role in pathophysiological mechanisms and/or could potentially be useful as a biomarker for ALVR.
Materials and methods
Study population
This study was conducted as a multi-center prospective study between June 2015 and 2016. Patients (≥ 18 years of age) who were admitted to the hospital with STEMI for the first time ever, and who underwent primary percutaneous coronary intervention (pPCI) within 12 h after onset of chest pain were evaluated. The definition of STEMI was according to the 3rd universal definition of MI [15] and was managed according to the European Society of Cardiology guidelines [16]. Inclusion and exclusion criteria are shown in Table 1. A total of ten patients were included in the study.
Study protocol
Clinical, demographic, laboratory and radiological findings were timely recorded in patient files during follow-up. After inclusion, a follow-up cardiac magnetic resonance (CMR) imaging was performed at baseline (2 weeks) and 6 months after index event. The assessment of miRNAs expression and was conducted at baseline (1 day), 2 and 6 weeks after index event. All blood samples were collected and immediately processed at similar timepoints (08:00 and 12:00 A.M.) to prevent, if any, circadian rhythm-associated variation in miRNA expression. Serum and plasma samples were stored at − 80 °C until assayed. After collecting the serum specimens of the whole sample, parameters were measured by the same device, in the same session and by the same laboratory staff in Tissue Typing Laboratory and Genetic Diagnosis Center of Ankara Diskapi Training and Research Hospital.
Ethical approval and informed consent
The study was executed according to the principles of the Declaration of Helsinki 2013 and approved by the local ethics committee at each participating center (Decision Date, No: 06.2013, 2013/106). Written informed consent was obtained from all patients.
Biochemical parameters
Venous blood samples were taken from the antecubital vein at the 24th h of the percutaneous intervention. Afterward, the blood sample was centrifuged at 1500 rpm for 10 min to measure the determined parameters. Total cholesterol was measured on the Hitachi Modular P800 (Roche Diagnostics Corp. Indiana, Indianapolis, USA) autoanalyser by a homogeneous enzymatic colorimetric method. Low-density lipoprotein cholesterol was calculated using the Friedewald method [17].
Analysis and relative quantification of miRNA
Selection of miRNAs is based on a database of disease-associated SNPs and microRNA target sites on 3′UTRs of human genes (http://mirdsnp.ccr.buffalo.edu/search.php# and http://www.mir2disease.org) and literature before 2016 (Supplementary Table 1). Exosomes were precipitated from the plasma miRCURY™ Exosome Isolation Kit—serum and plasma (Exiqon, Copenhagen, Denmark). Total RNA was extracted from the exosome pellet using the miRCURY™ RNA isolation kit-biofluids. The isolation protocol was carried out in accordance with the manufacturer’s directives. The exosomes were transferred to a new microcentrifuge tube and 60 μL of Lysis solution BF containing 16.67 μg/mL of MS2 bacteriophage RNA and RNA spike-in template mixture was added to the sample. The tube was mixed and incubated for 3 min at room temperature, followed by addition of 20 μL Protein Precipitation solution BF. The tube was mixed, incubated for 1 min at room temperature and centrifuged at 11,000×g for 3 min. The clear supernatant was transferred to a new collection tube, and 270 μL isopropanol was added. The solutions were mixed and transfer to a binding column. The column was incubated for 2 min at room temperature, and emptied using a vacuum-manifold. After 100 μL wash solution 1 Biwa’s added to the spin columns. The liquid was removed using a vacuum-manifold, and 700 μL wash solution 2 BF was added. The liquid was removed using a vacuum-manifold. To dry the columns entirely, the vacuum was extended to 2 min. The miRNAs were eluted in 50 µL RNase-free H2O and the purified miRNAs were stored at − 80 °C.
cDNA synthesis performed using the miRCURY LNA™ Universal Real-time (RT) microRNA cDNA synthesis kit (Exiqon). 7 μL RNA was reverse transcribed in 35 μL reactions using the miRCURY LNA™ Universal RT microRNA PCR, Polyadenylation and cDNA synthesis kit (Exiqon). cDNA was diluted 50× and assayed in 10 μL PCR reactions according to the protocol for miRCURY LNA™ Universal RT microRNA PCR; each microRNA was assayed once by qPCR on the microRNA Ready-to-Use PCR, Serum/Plasma Focus panel using ExiLENT SYBR® Green master mix. Negative controls excluding template from the reverse transcription reaction was performed and profiled like the samples. The amplification was performed in a LightCycler® 480 Real-Time PCR System (Roche) in 384 well plates. The amplification efficiency was calculated using algorithms similar to the LinReg software. Furthermore, assays must be detected with Ct < 37 to be included in the data analysis. All results were expressed as Ct and normalized to the calculated mean Ct of each sample (ΔCt). The relative expression was calculated using the comparative Ct method (2−ΔΔCt). miRNAs with values below the limit of detection were not included in the analysis.
Cardiac magnetic resonance imaging
Cardiac magnetic resonance imaging was performed using a 3-T scanner (Magnetom Skyra, Siemens Medical Systems, Erlangen, Germany) 2 weeks and 6 month, and the imaging protocol entailed acquiring one four-chamber view, cine short-axis sections (10-mm intervals for slice thickness of 6 mm), and one two-chamber view. Left ventricular (LV) systolic function indices were evaluated with the retrospective electrocardiogram-gated turbo-fast low angle shot (turbo-FLASH) sequence. Acquisition settings were: echo time 1.42 ms, repetition time 39 ms, flip angle 57°, voxel size 1.67 × 1.67 × 6 mm. LV function and volumes were measured using Syngo.via imaging software (Siemens Healthineers, Erlangen, Germany). LVEDV and LVESV were calculated with short-axis based planimetry from basal to apical level. Stroke volume was calculated as LVEDV minus LVESV, and LV ejection fraction (LVEF) was calculated as follows: LVEF = [(LVEDV − LVESV)/LVEDV] × 100. The definition of ALVR was a ≥ 13% increase in LVEDV and LVESV at the 6 months after AMI [18].
Statistical analysis
Statistical evaluation was performed using the Statistical Package for Social Sciences (SPSS) for Windows 20 (IBM SPSS Inc., Chicago, IL) program. The normal distribution of the data was evaluated with the Shapiro–Wilk test. Normally distributed numerical variables were shown as mean ± standard deviation, while numerical variables not showing normal distribution were shown as median (minimum, maximum). Categorical variables were expressed as numbers and percentages. Chi-Square, and Yates’ correction and Fisher’s Exact test were used for comparison of categorical data. Student T-test or Mann–Whitney U test was used to compare numerical variables between two groups according to the distribution of normality. miRNA and CMR levels in post-MI periods were evaluated using mixed model for repeated measurements. p < 0.05 (*) value was considered significant in statistical analysis.
Results
The study consisted of 10 male patients with a mean age of the cohort was 51.6 ± 6.0 (43–60) years. Four (40%) patients had a history of smoking, 2 (20%) patients had hypertension and 1 patient (10%) had diabetes mellitus. When compared with baseline LV dimensions, ALVR evaluated by CMR was detected in 5 (50%) patients in the 6th month. There was no significant difference in baseline demographic and laboratory findings in patients with ALVR compared to RLVR (Table 2).
At the 2 weeks after AMI evaluation by CMR, the mean EDV, median ESV, and mean LVEF (%) levels were not significantly different between patients with ALVR and RLVR. However, at the 6 months after AMI in ALVR group, mean EDV, and median ESV, were increased, mean LVEF (%) was decreased (Table 3).
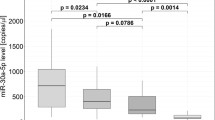
A panel of 178 miRNAs (Human Serum & Plasma miScript miRNA PCR Array) was profiled in ten patients (Supplement Table 2). In all candidate exosomes, 8 miRNAs (miR-199a-5p, miR-23b-3p, miR-26b-5p, miR-301a-3p, miR-374a-5p, miR-423-5p, miR-483-5p and miR-652-3p) were associated with ALVR during the follow-up period (Table 4) (Fig. 1). According to this; at the 1st day after AMI, miR-199a-5p (fold change: 0.30; p = 0.050), miR-23b-3p (fold change: 0.56; p = 0.050), and miR-483-5p (fold change: 0.34; p = 0.034) were significantly downregulated and miR-26b-5p (fold change: 2.35; p = 0.028) and miR-652-3p (fold change: 1.79; p = 0.050) were significantly upregulated in ALVR compared to RLVR groups (Fig. 2). At the 2 weeks after AMI, miR-374a-5p (fold change: 0.28; p = 0.050) was significantly downregulated in ALVR compared to RLVR groups. At the 6 weeks after AMI, miR-301a-3p (fold change: 0.24; p = 0.050), and miR-374a-5p (fold change: 0.48; p = 0.050) were significantly downregulated and miR-423-5p (fold change: 1.76; p = 0.047) was significantly upregulated ALVR compared to RLVR group (Fig. 3). The fold changes of miRNAs in ALVR versus RLVR group are shown in Table 4.
Discussion
The current study was the first to assess exosomal miRNA expression in more than one period in patients with AMI. We determined that 8 miRNAs from the panel of 178 exosomal miRNA were significantly up- or downregulated in patients with ALVR (miR-199a-5p, miR-23b-3p, miR-26b-5p, miR-301a-3p, miR-374a-5p, miR-423-5p, miR-483-5p, miR-652-3p). The vast majority of these miRNAs differed significantly on the 1st day after AMI. Furthermore, in serial measurements, only miR-374a-5p was downregulated on the 2 and 6 weeks post-MI. MiR-301a-3p was downregulated on the 6 weeks post-MI, and miR-423-5p was upregulated on the 6 weeks post-MI. Serial measurements gave information about the period in which miRNAs play a role in cardiac remodeling.
The stability of circulating miRNAs and their regulation in pathological conditions has sparked interest in their use as biomarkers. The heart reacts to hemodynamic overload and injury by stimulating myocyte hypertrophy, reprogramming of gene expression and remodeling of extracellular matrix [19]. In myocardial infarction pathology, miRNAs are involved in the regulation of different types of cell death, including necrosis and apoptosis [20, 21]. Increasing evidences have shown an important role for microRNA (miRNA)-regulated mechanisms in myocardial healing or ALVR after AMI [22,23,24,25]. Overexpression of some miRNAs (miR-133a-3p, miR-1a-3p, miR-27b-3p and miR-208a-3p) preserves cardiac functions, while inhibition has been associated with hypertrophy and dysfunction. On the other hand, some miRNAs (MiR-21-5p, miR-378-5p and miR-223-3p) were found to be overexpressed in heart failure patients [26]. However, there may be as yet unidentified miRNAs that may play a role in both MI and ALVR.
MiR-199a-5p has been shown to be abundant in the myocardium and its downregulation plays an important role in the prevention of cardiac hypertrophy and heart failure [27]. It is reported that miR-199a-5p rapidly decreases in cardiomyocytes exposed to hypoxia [28]. In studies performed in different heart failure models, it has been shown that miR-199a-5p upregulation prevents cell death [29, 30]. In contrast, other studies have been shown that high expression of miR-199a-5p after MI contributes to heart failure development [31,32,33]. We found that baseline expression of miR-199a-5p was downregulated in ALVR group, while there was no difference in the miR-199a-5p expression between ALVR and RLVR groups in the later periods. Low expression of miR-199a-5p may play a role in ALVR with potential mechanisms such as apoptosis, pathological cardiac myocyte hypertrophy and cardiac homeostasis due to inflammatory response [28, 34,35,36].
MiR-23b-3p, which plays a role in monocyte/macrophage inflammatory reaction, is upregulated under cardiac stress conditions [37, 38]. Zhang et al. [39] showed that miR-23b-3p was upregulated at the time of acute MI and decreased after percutaneous coronary intervention. This may be due to the post-MI period miR-23b-3p is associated with migration of phagocytes, proliferation of mononuclear leukocytes and cell movement of smooth muscle cells [40]. In our study, baseline expression of miR-23b-3p was downregulated in the ALVR group. These findings suggest that downregulation of miR-23b-3p may be associated with an unfavorable inflammatory response. On the other hand, the excessive expression of miR-26b-5p, which plays a role in the development of cardiac hypertrophy in physiological conditions possible as an outcome of blocking the initiation of autophagy, has been found to be associated with increased cardiovascular events in patients with AMI [41, 42]. This hypothesis is consistent with the upregulation of miR-26b-5p expression in our study. Previous studies have shown that miR-26b-5p is upregulated in patients with heart failure [43, 44].
MiR-483-5p, which plays a role in the regulation of apoptosis and are upregulated during the acute MI process, were found at high levels in our study, especially in patients with RLVR. This role may be due to the triggering of IGF2 by the presence of miR-483-5p upregulation and thus the involvement of pro-inflammatory mechanisms in the regulation of NF-kB- and interleukin-6 mediated pathways [45]. To the best of our knowledge, we show for the first time in this study that downregulation of MiR-483-5p expression is associated with ALVR. Therefore, the role of miR-483-5p in LV remodeling requires further research, examining host gene transcription and protein expression rather than a biomarker.
In the early phase of AMI, the upregulation of MiR-652-3p expression was associated with ALVR. Huang et al. [46] found that the inhibition of miR-652-3p promotes cyclin D2 expression, increasing endothelial repair and decreasing atherosclerosis. Wang et al. [47] showed that an anaerobic condition triggers the accumulation of fragmented mitochondria and leads to apoptotic cell death by a mechanism of MFACR-dependent inhibition of miR-652-3p and uninterrupted expression of the MTP18 protein.
We found that majority of ALVR-associated miRNAs developed significant responses in the early phase AMI. However, in the follow-up period, differences were found in the expression of miR-423-5p, miR-301a-3p, and miR-374a-5p. It is known that the level of circulating miR-423-5p is a useful biomarker in demonstrating LV remodeling after AMI [48]. Bauters et al. [49] found that an increase in miR-423-5p levels over periods compared to baseline post-MI. Consistently, in the current study, it was observed that the miR-423-5p expression at the 6 weeks post-MI were higher in the patients with ALVR. MiR-301a, second late onset miRNA, regulates calsarcin-1 and cofilin-2, which have roles in the development of cardiomyopathy [50]. In addition, miR-374a-5p may have protective effects against myocardial ischemia–reperfusion injury and can be expected to be upregulated in the early phase of acute MI due to percutaneous coronary intervention [51]. It seems consistent that the miR-301a-3p level, which has been shown to be indirectly related to heart failure, showed a significant difference at 6 weeks post-MI, but it was expected that miR-374a-5p would show an increased response in the early period of AMI. Therefore, more evidence is needed to understand the effect of miRNAs in ALVR after AMI.
Cardiac remodeling can occur weeks to months after reperfusion. Exosomal miRNA is known to show extra stability, even under different storage conditions. Therefore, post-AMI exosome miRNAs may be good biomarkers based on their stability under various storage conditions, with future diagnostic and therapeutic uses. Although clinical indicators or circulating biomarkers are helpful in determining the risk of ALVR after AMI, but they have certain limitations. Increasing evidence indicates that miRNAs expressions may be an important biomarker in detecting high-risk patients post-MI. Besides, miRNAs may be a new individualized targeted therapy on adverse LV remodeling. MiRNA-based therapies will have the ability to modulate multiple target genes or networks involved in the cardiac remodeling process [52]. The regulation of apoptotic pathway could represent a therapeutic approach to treat apoptotic-related cardiac disease [53]. Thus, normalizing miRNA expression in the heart represents a new approach to the pharmacotherapy of heart disease.
There are some important limitations of our study. It was recognized that this clinical study sample was small with limited power for full adjustment of clinical and RNA-based biomarkers. On the other hand, we could not conduct experiments to modulate miRNA levels in native human tissues. However, this was designed as a small-sized preliminary study rather than one to determine a cause-effect relationship. Myocardial healing processes comprise complex interactions that involve many molecules. Hence, eliminating individual variations and interactions are almost impossible. In addition, our entire study population was male. MiRNA transcriptome are associated with significant gender differences that may affect cardiac homeostasis [54, 55].
As a conclusion; it was observed that certain exosomal miRNAs are associated with ALVR after AMI. It was also found that the majority of miRNAs showed significant changes in the early period of AMI, while the remaining ones showed changes over time. The detection of miRNAs with, different roles in various pathways might provide insight for ALVR after AMI. Investigating the roles of miRNAs in modulating various aspects of the LV remodeling process will contribute to identifying the potential implications of miRNA biology in the field of heart failure. However, larger confirmatory studies to investigate the miRNA’s of interest are needed and pre-clinical experiments are necessary to understand their mechanistic role in the development of ALVR.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on request.
References
Azevedo PS, Polegato BF, Minicucci MF, Paiva SA, Zornoff LA (2016) Cardiac remodeling: concepts, clinical impact, pathophysiological mechanisms and pharmacologic treatment. Arq Bras Cardiol 106:62–69. https://doi.org/10.5935/abc.20160005
Piek A, de Boer RA, Sillje HH (2016) The fibrosis-cell death axis in heart failure. Heart Fail Rev 21:199–211. https://doi.org/10.1007/s10741-016-9536-9
Bhatt AS, Ambrosy AP, Velazquez EJ (2017) Adverse remodeling and reverse remodeling after myocardial infarction. Curr Cardiol Rep 19:71. https://doi.org/10.1007/s11886-017-0876-4
Cruz MS, da Silva AMG, de Souza KSC, Luchessi AD, Silbiger VN (2020) miRNAs emerge as circulating biomarkers of post-myocardial infarction heart failure. Heart Fail Rev 25:321–329. https://doi.org/10.1007/s10741-019-09821-1
Thum T, Condorelli G (2015) Long noncoding RNAs and microRNAs in cardiovascular pathophysiology. Circ Res 116:751–762. https://doi.org/10.1161/CIRCRESAHA.116.303549
Melman YF, Shah R, Das S (2014) MicroRNAs in heart failure: is the picture becoming less miRky? Circ Heart Fail 7:203–214. https://doi.org/10.1161/CIRCHEARTFAILURE.113.000266
Fiedler J, Jazbutyte V, Kirchmaier BC, Gupta SK, Lorenzen J, Hartmann D, Galuppo P, Kneitz S, Pena JT, Sohn-Lee C, Loyer X, Soutschek J, Brand T, Tuschl T, Heineke J, Martin U, Schulte-Merker S, Ertl G, Engelhardt S, Bauersachs J, Thum T (2011) MicroRNA-24 regulates vascularity after myocardial infarction. Circulation 124:720–730. https://doi.org/10.1161/CIRCULATIONAHA.111.039008
Lakhani HV, Khanal T, Gabi A, Yousef G, Alam MB, Sharma D, Aljoudi H, Puri N, Thompson E, Shapiro JI, Sodhi K (2018) Developing a panel of biomarkers and miRNA in patients with myocardial infarction for early intervention strategies of heart failure in West Virginian population. PLoS ONE 13:e0205329. https://doi.org/10.1371/journal.pone.0205329
Zile MR, Mehurg SM, Arroyo JE, Stroud RE, DeSantis SM, Spinale FG (2011) Relationship between the temporal profile of plasma microRNA and left ventricular remodeling in patients after myocardial infarction. Circ Cardiovasc Genet 4:614–619. https://doi.org/10.1161/CIRCGENETICS.111.959841
Divakaran V, Mann DL (2008) The emerging role of microRNAs in cardiac remodeling and heart failure. Circ Res 103:1072–1083. https://doi.org/10.1161/CIRCRESAHA.108.183087
Lv P, Zhou M, He J, Meng W, Ma X, Dong S, Meng X, Zhao X, Wang X, He F (2014) Circulating miR-208b and miR-34a are associated with left ventricular remodeling after acute myocardial infarction. Int J Mol Sci 15:5774–5788. https://doi.org/10.3390/ijms15045774
Zhang L, Ding H, Zhang Y, Wang Y, Zhu W, Li P (2020) Circulating MicroRNAs: biogenesis and clinical significance in acute myocardial infarction. Front Physiol 11:1088. https://doi.org/10.3389/fphys.2020.01088
Cheng L, Sharples RA, Scicluna BJ, Hill AF (2014) Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J Extracell Vesicles. https://doi.org/10.3402/jev.v3.23743
Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO (2007) Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9:654–659. https://doi.org/10.1038/ncb1596
Thygesen K, Alpert JS, Jaffe AS, Simoons ML, Chaitman BR, White HD, Writing Group on the Joint ESC/ACCF/AHA/WHF Task Force for the Universal Definition of Myocardial Infarction, Thygesen K, Alpert JS, White HD, Jaffe AS, Katus HA, Apple FS, Lindahl B, Morrow DA, Chaitman BA, Clemmensen PM, Johanson P, Hod H, Underwood R, Bax JJ, Bonow RO, Pinto F, Gibbons RJ, Fox KA, Atar D, Newby LK, Galvani M, Hamm CW, Uretsky BF, Steg PG, Wijns W, Bassand JP, Menasche P, Ravkilde J, Ohman EM, Antman EM, Wallentin LC, Armstrong PW, Simoons ML, Januzzi JL, Nieminen MS, Gheorghiade M, Filippatos G, Luepker RV, Fortmann SP, Rosamond WD, Levy D, Wood D, Smith SC, Hu D, Lopez-Sendon JL, Robertson RM, Weaver D, Tendera M, Bove AA, Parkhomenko AN, Vasilieva EJ, Mendis S, ESC Committee for Practice Guidelines (CPG) (2012) Third universal definition of myocardial infarction. Eur Heart J 33:2551–2567. https://doi.org/10.1093/eurheartj/ehs184
Ibanez B, James S, Agewall S, Antunes MJ, Bucciarelli-Ducci C, Bueno H, Caforio ALP, Crea F, Goudevenos JA, Halvorsen S, Hindricks G, Kastrati A, Lenzen MJ, Prescott E, Roffi M, Valgimigli M, Varenhorst C, Vranckx P, Widimsky P, Group ESCSD (2018) 2017 ESC guidelines for the management of acute myocardial infarction in patients presenting with ST-segment elevation: the Task Force for the management of acute myocardial infarction in patients presenting with ST-segment elevation of the European Society of Cardiology (ESC). Eur Heart J 39:119–177. https://doi.org/10.1093/eurheartj/ehx393
Friedewald WT, Levy RI, Fredrickson DS (1972) Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem 18:499–502
Bulluck H, Go YY, Crimi G, Ludman AJ, Rosmini S, Abdel-Gadir A, Bhuva AN, Treibel TA, Fontana M, Pica S, Raineri C, Sirker A, Herrey AS, Manisty C, Groves A, Moon JC, Hausenloy DJ (2017) Defining left ventricular remodeling following acute ST-segment elevation myocardial infarction using cardiovascular magnetic resonance. J Cardiovasc Magn Reson 19:26. https://doi.org/10.1186/s12968-017-0343-9
Small EM, Frost RJ, Olson EN (2010) MicroRNAs add a new dimension to cardiovascular disease. Circulation 121:1022–1032. https://doi.org/10.1161/CIRCULATIONAHA.109.889048
Wang JX, Zhang XJ, Li Q, Wang K, Wang Y, Jiao JQ, Feng C, Teng S, Zhou LY, Gong Y, Zhou ZX, Liu J, Wang JL, Li PF (2015) MicroRNA-103/107 regulate programmed necrosis and myocardial ischemia/reperfusion injury through targeting FADD. Circ Res 117:352–363. https://doi.org/10.1161/CIRCRESAHA.117.305781
Higashi K, Yamada Y, Minatoguchi S, Baba S, Iwasa M, Kanamori H, Kawasaki M, Nishigaki K, Takemura G, Kumazaki M, Akao Y, Minatoguchi S (2015) MicroRNA-145 repairs infarcted myocardium by accelerating cardiomyocyte autophagy. Am J Physiol Heart Circ Physiol 309:H1813–H1826. https://doi.org/10.1152/ajpheart.00709.2014
van Boven N, Kardys I, van Vark LC, Akkerhuis KM, de Ronde MWJ, Khan MAF, Merkus D, Liu Z, Voors AA, Asselbergs FW, van den Bos EJ, Boersma E, Hillege H, Duncker DJ, Pinto YM, Postmus D (2018) Serially measured circulating microRNAs and adverse clinical outcomes in patients with acute heart failure. Eur J Heart Fail 20:89–96. https://doi.org/10.1002/ejhf.950
de Gonzalo-Calvo D, Cediel G, Bar C, Nunez J, Revuelta-Lopez E, Gavara J, Rios-Navarro C, Llorente-Cortes V, Bodi V, Thum T, Bayes-Genis A (2018) Circulating miR-1254 predicts ventricular remodeling in patients with ST-segment-elevation myocardial infarction: a cardiovascular magnetic resonance study. Sci Rep 8:15115. https://doi.org/10.1038/s41598-018-33491-y
Tikhomirov R, Donnell BR, Catapano F, Faggian G, Gorelik J, Martelli F, Emanueli C (2020) Exosomes: from potential culprits to new therapeutic promise in the setting of cardiac fibrosis. Cells. https://doi.org/10.3390/cells9030592
Chen C, Ponnusamy M, Liu C, Gao J, Wang K, Li P (2017) MicroRNA as a therapeutic target in cardiac remodeling. Biomed Res Int 2017:1278436. https://doi.org/10.1155/2017/1278436
Peters LJF, Biessen EAL, Hohl M, Weber C, van der Vorst EPC, Santovito D (2020) Small things matter: relevance of MicroRNAs in cardiovascular disease. Front Physiol 11:793. https://doi.org/10.3389/fphys.2020.00793
Song XW, Li Q, Lin L, Wang XC, Li DF, Wang GK, Ren AJ, Wang YR, Qin YW, Yuan WJ, Jing Q (2010) MicroRNAs are dynamically regulated in hypertrophic hearts, and miR-199a is essential for the maintenance of cell size in cardiomyocytes. J Cell Physiol 225:437–443. https://doi.org/10.1002/jcp.22217
Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, Vatner DE, Vatner SF, Abdellatif M (2009) Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res 104:879–886. https://doi.org/10.1161/CIRCRESAHA.108.193102
Abdellatif M (2011) Cardioprotective microRNAs. Pediatr Cardiol 32:311–316. https://doi.org/10.1007/s00246-010-9882-7
el Azzouzi H, Leptidis S, Dirkx E, Hoeks J, van Bree B, Brand K, McClellan EA, Poels E, Sluimer JC, van den Hoogenhof MM, Armand AS, Yin X, Langley S, Bourajjaj M, Olieslagers S, Krishnan J, Vooijs M, Kurihara H, Stubbs A, Pinto YM, Krek W, Mayr M, da Costa Martins PA, Schrauwen P, De Windt LJ (2013) The hypoxia-inducible microRNA cluster miR-199a approximately 214 targets myocardial PPARdelta and impairs mitochondrial fatty acid oxidation. Cell Metab 18:341–354. https://doi.org/10.1016/j.cmet.2013.08.009
van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN (2006) A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc Natl Acad Sci USA 103:18255–18260. https://doi.org/10.1073/pnas.0608791103
Haghikia A, Missol-Kolka E, Tsikas D, Venturini L, Brundiers S, Castoldi M, Muckenthaler MU, Eder M, Stapel B, Thum T, Haghikia A, Petrasch-Parwez E, Drexler H, Hilfiker-Kleiner D, Scherr M (2011) Signal transducer and activator of transcription 3-mediated regulation of miR-199a-5p links cardiomyocyte and endothelial cell function in the heart: a key role for ubiquitin-conjugating enzymes. Eur Heart J 32:1287–1297. https://doi.org/10.1093/eurheartj/ehq369
Liu X, Meng H, Jiang C, Yang S, Cui F, Yang P (2016) Differential microRNA expression and regulation in the rat model of post-infarction heart failure. PLoS ONE 11:e0160920. https://doi.org/10.1371/journal.pone.0160920
Huang H, Xie S, Gu X, Xiang B, Zhong Z, Huang P, Gao Y, Li P (2021) Higher circulating miR-199a-5p indicates poor aerobic exercise capacity and associates with cardiovascular dysfunction during chronic exposure to high altitude. Front Physiol 12:587241. https://doi.org/10.3389/fphys.2021.587241
Duygu B, Poels EM, Juni R, Bitsch N, Ottaviani L, Olieslagers S, de Windt LJ, da Costa Martins PA (2017) miR-199b-5p is a regulator of left ventricular remodeling following myocardial infarction. Noncoding RNA Res 2:18–26. https://doi.org/10.1016/j.ncrna.2016.12.002
Yan M, Yang S, Meng F, Zhao Z, Tian Z, Yang P (2018) MicroRNA 199a-5p induces apoptosis by targeting JunB. Sci Rep 8:6699. https://doi.org/10.1038/s41598-018-24932-9
Wang H, Cai J (2017) The role of microRNAs in heart failure. Biochim Biophys Acta 1863:2019–2030. https://doi.org/10.1016/j.bbadis.2016.11.034
Yang Z, Guo Z, Dong J, Sheng S, Wang Y, Yu L, Wang H, Tang L (2018) miR-374a regulates inflammatory response in diabetic nephropathy by targeting MCP-1 expression. Front Pharmacol 9:900. https://doi.org/10.3389/fphar.2018.00900
Zhang J, Li Y, Zhao Q (2018) Circulating miR-23b as a novel biomarker for early risk stratification after ST-elevation myocardial infarction. Med Sci Monit 24:1517–1523. https://doi.org/10.12659/msm.908060
Jiang Y, Zhang M, He H, Chen J, Zeng H, Li J, Duan R (2013) MicroRNA/mRNA profiling and regulatory network of intracranial aneurysm. BMC Med Genomics 6:36. https://doi.org/10.1186/1755-8794-6-36
John Clotaire DZ, Zhang B, Wei N, Gao R, Zhao F, Wang Y, Lei M, Huang W (2016) MiR-26b inhibits autophagy by targeting ULK2 in prostate cancer cells. Biochem Biophys Res Commun 472:194–200. https://doi.org/10.1016/j.bbrc.2016.02.093
Han M, Yang Z, Sayed D, He M, Gao S, Lin L, Yoon S, Abdellatif M (2012) GATA4 expression is primarily regulated via a miR-26b-dependent post-transcriptional mechanism during cardiac hypertrophy. Cardiovasc Res 93:645–654. https://doi.org/10.1093/cvr/cvs001
Marfella R, Di Filippo C, Potenza N, Sardu C, Rizzo MR, Siniscalchi M, Musacchio E, Barbieri M, Mauro C, Mosca N, Solimene F, Mottola MT, Russo A, Rossi F, Paolisso G, D’Amico M (2013) Circulating microRNA changes in heart failure patients treated with cardiac resynchronization therapy: responders vs. non-responders. Eur J Heart Fail 15:1277–1288. https://doi.org/10.1093/eurjhf/hft088
Ovchinnikova ES, Schmitter D, Vegter EL, Ter Maaten JM, Valente MA, Liu LC, van der Harst P, Pinto YM, de Boer RA, Meyer S, Teerlink JR, O’Connor CM, Metra M, Davison BA, Bloomfield DM, Cotter G, Cleland JG, Mebazaa A, Laribi S, Givertz MM, Ponikowski P, van der Meer P, van Veldhuisen DJ, Voors AA, Berezikov E (2016) Signature of circulating microRNAs in patients with acute heart failure. Eur J Heart Fail 18:414–423. https://doi.org/10.1002/ejhf.332
Harling L, Lambert J, Ashrafian H, Darzi A, Gooderham NJ, Athanasiou T (2017) Elevated serum microRNA 483-5p levels may predict patients at risk of post-operative atrial fibrillation. Eur J Cardiothorac Surg 51:73–78. https://doi.org/10.1093/ejcts/ezw245
Huang R, Hu Z, Cao Y, Li H, Zhang H, Su W, Xu Y, Liang L, Melgiri ND, Jiang L (2019) MiR-652-3p inhibition enhances endothelial repair and reduces atherosclerosis by promoting Cyclin D2 expression. EBioMedicine 40:685–694. https://doi.org/10.1016/j.ebiom.2019.01.032
Wang K, Gan TY, Li N, Liu CY, Zhou LY, Gao JN, Chen C, Yan KW, Ponnusamy M, Zhang YH, Li PF (2017) Circular RNA mediates cardiomyocyte death via miRNA-dependent upregulation of MTP18 expression. Cell Death Differ 24:1111–1120. https://doi.org/10.1038/cdd.2017.61
Tijsen AJ, Creemers EE, Moerland PD, de Windt LJ, van der Wal AC, Kok WE, Pinto YM (2010) MiR423-5p as a circulating biomarker for heart failure. Circ Res 106:1035–1039. https://doi.org/10.1161/CIRCRESAHA.110.218297
Bauters C, Kumarswamy R, Holzmann A, Bretthauer J, Anker SD, Pinet F, Thum T (2013) Circulating miR-133a and miR-423-5p fail as biomarkers for left ventricular remodeling after myocardial infarction. Int J Cardiol 168:1837–1840. https://doi.org/10.1016/j.ijcard.2012.12.074
Rangrez AY, Hoppe P, Kuhn C, Zille E, Frank J, Frey N, Frank D (2017) MicroRNA miR-301a is a novel cardiac regulator of Cofilin-2. PLoS ONE 12:e0183901. https://doi.org/10.1371/journal.pone.0183901
Huang ZQ, Xu W, Wu JL, Lu X, Chen XM (2019) MicroRNA-374a protects against myocardial ischemia–reperfusion injury in mice by targeting the MAPK6 pathway. Life Sci 232:116619. https://doi.org/10.1016/j.lfs.2019.116619
van Balkom BW, de Jong OG, Smits M, Brummelman J, den Ouden K, de Bree PM, van Eijndhoven MA, Pegtel DM, Stoorvogel W, Wurdinger T, Verhaar MC (2013) Endothelial cells require miR-214 to secrete exosomes that suppress senescence and induce angiogenesis in human and mouse endothelial cells. Blood. https://doi.org/10.1182/blood-2013-02-478925
Ren XP, Wu J, Wang X, Sartor MA, Jones K, Qian J, Nicolaou P, Pritchard TJ, Fan GC (2009) MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20. Circulation 119:2357–2366. https://doi.org/10.1161/CIRCULATIONAHA.108.814145
Tsuji M, Kawasaki T, Matsuda T, Arai T, Gojo S, Takeuchi JK (2017) Sexual dimorphisms of mRNA and miRNA in human/murine heart disease. PLoS ONE 12:e0177988. https://doi.org/10.1371/journal.pone.0177988
Florijn BW, Bijkerk R, van der Veer EP, van Zonneveld AJ (2018) Gender and cardiovascular disease: are sex-biased microRNA networks a driving force behind heart failure with preserved ejection fraction in women? Cardiovasc Res 114:210–225. https://doi.org/10.1093/cvr/cvx223
Funding
The authors declared that this research received financial support from the Ministry of Health of the Republic of Turkey 2015/SAGEM-2/001 project.
Author information
Authors and Affiliations
Contributions
Concept—FE, NE; Design—EF, NE; Supervision—FE; Data collection &/or processing—FE, KE, EK, IBU, AK and NE; Analysis &/or interpretation—FE, KE, EK, IBU and AK; Literature search—FE, KE, EK, IBU, AK and NE; Writing—FE; Critical review—NE.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare they have no conflicts of interest.
Ethical approval
The study was performed in accordance with the Declaration of Helsinki, and approved by the Faculty Of Medicine Non-Drug Clinical Research Ethics Committee of the Ankara Yildirim Beyazit University, on 24 June 2013, under Decision No. 2013/106.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Eyyupkoca, F., Ercan, K., Kiziltunc, E. et al. Determination of microRNAs associated with adverse left ventricular remodeling after myocardial infarction. Mol Cell Biochem 477, 781–791 (2022). https://doi.org/10.1007/s11010-021-04330-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-021-04330-y