Abstract
In previous studies, long non-coding RNA LINC01410 (LINC01410) has been found to promote cells proliferation and invasion in colon and gastric cancers. However, the function of LINC01410 in endometrial cancer (EC) is still elusive. The expression patterns of LINC01410/miR-23c/Chromodomain Helicase DNA-Binding Protein 7 (CHD7) in EC tissues and the prognosis of patients with different expression of LINC01410/miR-23c/CHD7 were determined by consulting TCGA database. EC patients with complete clinical data were applied for clinicopathological correlation analysis. The biological characteristics of EC cells were analyzed with the support of CCK-8 and transwell assays. CHD7 expression was assessed by qRT-PCR and western blot assays. Targeted associations between LINC01410 and miR-23c, as well as miR-23c and CHD7 were speculated by prediction website and verified by dual-luciferase assay. Rescue assays were performed to explore the interrelation among LINC01410, miR-23c and CHD7. Our data illustrated that LINC01410 high expression was presented in EC tissues and was positively related to the poor prognosis of patients in EC, as well as the malignant behaviors of EC cells. Through bioinformatics analysis, we surmised that LINC01410/miR-23c/CHD7 may play a role through the formation of competing endogenous RNA (ceRNA) mechanism. CHD7 expression was positively regulated by LINC01410, and inversely controlled by miR-23c. Furthermore, the promoting effects of miR-23c inhibitor or CHD7 upregulation on EC cell growth and aggressiveness were attenuated by LINC01410 silencing. Our results indicated that high expression of LINC01410 promoted EC cell progression through modulating miR-23c/CHD7 axis, providing a new direction for revealing the molecular mechanism of EC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Endometrial cancer (EC) is not only one of the most common gynecological malignancies but also the most common cause of hysterectomy [1, 2]. Currently, the incidence of EC and mortality from EC has gone up, which may be connected with more common obesity and metabolic syndrome [3]. Around the world, about 142,000 women develop EC each year, and 42,000 women die from it [4]. The prognosis is still worse in patients with EC at the advanced stage or with a high risk of recurrence [4]. Therefore, it is essential to strengthen the understanding of the molecular mechanism of EC pathogenesis, to identify new therapeutic targets, and to develop more effective EC treatment strategies.
Long non-coding RNAs (lncRNAs), which belong to the family of non-coding RNAs (ncRNAs), account for more than 80% of human genomic transcripts [5]. A great amount of studies disclosed that lncRNAs possess crucial functions in numerous cellular processes, covering cell proliferation and cycle, as well as tumor metastasis [6, 7], through modulating the expression of protein-coding gene, altering epigenetic modulation, as well as serving as bait that sponging microRNAs (miRNAs) [8, 9]. Until now, several studies have indicated that plenty of lncRNAs, including HOTAIR, CCAT2, TDRG1, SNHG16, MEG3, were abnormally expressed in EC, and were involved in the development, metastasis, and drug resistance of EC cells [10,11,12,13,14]. However, study on LINC01410 in EC has not been well done. The reports about the function of LINC01410 in tumor were limited in gastric cancer, colon cancer, papillary thyroid carcinoma, and pancreatic cancer. For example, a earlier study has shown that LINC01410 promoted angiogenesis and metastasis of gastric cancer by reducing miR-532 expression [15]. Furthermore, it has been revealed that LINC01410 promoted cell growth and invasion in colon cancer via downregulating miR-3128 expression [16]. These findings insinuated that LINC01410 may play a crucial role in the proliferation and invasion of cancer cells. By enquiring TCGA database, LINC01410 showed a tendency of high expression, and its high expression was positively correlated with worse prognosis of patients with EC, suggesting that LINC01410 was associated with the progression of EC. In light of all this, we performed this study to investigate the function of LINC01410 in EC.
In 2011, the competing endogenous RNA (ceRNA) hypothesis was proposed by Salmena et al. which hold that any RNA molecules that harbor miRNA-response elements (MREs) can isolate miRNAs from other targets sharing the same MREs, thus regulating their functions [17]. Importantly, this assumption has been confirmed by some appended studies, which presented that lncRNAs could act as ceRNAs that contend with miRNA-binding sites, thus suppressing the expression of mRNAs that targeted by miRNA [18, 19]. Interestingly, the ceRNA mechanism has been revealed in several human tumors, including EC. For example, the lncRNA SNHG16 regulated glycolysis and proliferation of EC via miR-490-3p/HK2 axis [13], and lncRNA OGFRP1 promoted EC development through modulating miR-124-3p/SIRI1 axis [20]. Besides that, miRNAs executed crucial roles in numerous cancers. For example, a research from Muhammad et al. discovered that anti-miR-203 suppressed breast cancer growth and stemness by targeting SOCS3 [21]. Moreover, miR-23c inhibited tumor growth in hepatocellular carcinoma via targeting ERBB2IP [22]. All the researches illustrated the importance of miRNAs in cancers. By biological analysis, we preliminarily constructed “LINC01410/miR-23c/ Chromodomain Helicase DNA-Binding Protein 7 (CHD7)” network. However, the function of the network and its exact mechanisms in EC progression remain poorly understood.
During this study, we expounded the effects and underlying mechanism of LINC01410 in EC. We detected the expression of LINC01410 in EC samples and evaluated the function of LINC01410 aberrant expression on the biological characteristics of EC cells. More importantly, we explored the regulatory mechanisms between LINC01410 and miR-23c, as well as miR-23c and CHD7. All the consequences might afford a theoretical foundation for projecting new biomarkers for remedying EC.
Materials and methods
Bioinformatics analysis
Data from The Cancer Genome Atlas (TCGA) (https://cancergenome.nih.gov/) database including 552 tumor tissues and 35 normal tissues was employed to assess the expression of LINC01410 and CHD7, and conduct prognosis analysis. Data from TCGA including 546 tumor tissues and 33 normal tissues was applied to evaluate the expression of miR-23c and perform prognosis analysis. Patients’ information from TCGA with complete clinical data was used for correlation analysis. LncBase website was used to speculate the target miRNAs of LINC01410. Targetscan website was utilized to predict the target mRNAs of miR-23c.
Cell culture and treatment
Human EC cell lines containing RL95-2, HEC-1-A and KLE were purchased from ATCC (USA), and the normal uterine endometrial epithelial cells (NUEEC) were obtained from chiscientific.biomart.cn (Jiangyin, Jiangsu, China). Cells were incubated in RPMI-1640 medium appending with 10% fetal bovine serum, 100 U/mL penicillin, and 0.1 mg/mL streptomycin in 37 °C incubator with 5% CO2.
Sangon (Shanghai, China) provided the sequences of si-con (5′-CGAACTCACTGGTCTGACC-3′), si-LINC01410#1 (5′- TTCAAGAAATGGGAGATTCGACT-3′), si-LINC01410#2 (5′-TTGCTGATTGAGCAAGAATTAAA-3′), si-CHD7 (5′-CTAACGTACCTAACCTATTAA-3′), miR-23c mimic/inhibitor/NC/pcDNA3.1/pcDNA3.1-LINC01410/pcDNA3.1-CHD7. All of the sequences were transfected into cells using Lipofectamine2000 (11668-027, Invitrogen, USA) based on the manufacture’s instruction to down or upregulate LINC01410, miR-23c or CHD7 expression.
RNA extraction and quantitative real-time PCR (qRT-PCR)
We separated the whole RNA from the treated cells with the support of TRIzol reagent (15596-026, Invitrogen, USA). The quantity and purity of RNA were assessed using a SmartSpec Plus spectrophotometer (BioRad, USA). qPCR was executed to amplify the target genes using a qRT-PCR kit in a 7300HT real-time PCR system. To evaluate the expression of miRNA, qPCR was operated with the support of miScript II RT Kit and miScript SYBR Green PCR kit in a 7300HT real-time PCR system. GAPDH and U6 were regarded as the internal standards for mRNA and miRNA expression, severally. Relative quantification of genes expression was carried out using 2−ΔΔCt method. The primer sequences were presented as below:
LINC01410 F: 5′-GTGACA AGAATGGCCCAAGC-3′,
R: 5′-ACTGTGCACCTG TTACACCA-3′;
CHD7 F: 5′-CCTTTCCATGCTGAAGTTCCTGC-3′,
R: 5′-TCAGGCATACCGACTCGTTCCA-3′;
GAPDH F: 5′-TGTGTCCGTCGTGGATCTGA-3′,
R: 5′-CCTGCTTCACCACCTTCTTGA-3’.
Western blot
Cells from each group were lysed on ice in RIPA buffer, and the concentrations of each protein were assessed by utilizing BCA methods. After denaturing (5 min, 95 °C), the proteins were isolated using SDS-PAGE and transferred to a PVDF membrane. After blocking, the proteins on PVDF membranes were incubated with primary antibodies (Abcam, UK) covering CHD7 (ab176807, 1:5000), Cyclin D1 (ab16663, 1:200), CDK6 (ab124821, 1:50,000), Bcl2 (ab32124,1:1000), Bax (ab32503, 1:5000) and GAPDH (ab181602, 1:10,000) for 24 h at 4 °C. Afterwards, the proteins were incubated with secondary antibody for 2 h at about 25 °C. Finally, ECL was added to develop the signals, and Image Quant LAS 500 was applied to capture the image.
Cell proliferation assay
After 24-h post-transfection, the cells were implanted into 96-well plates with the density of 1000 cells/well for the following analysis with the support of Cell Counting Kit-8 (CCK-8, CK04, Japan). The cell viability was determined at 0 h, 24 h, 48 h and 72 h. Briefly, 15 μL of CCK-8 solution was filled into the well equipped with cells for a 1.5 h incubation at 37 °C. After shaking, the optical density at 450 nm was detected using a spectrophotometer (BioRad, USA).
Cell invasion and migration assays
Transwell assay was performed to estimate cell migration and invasion. For invasion assay, the transwell chambers were pre-coated with Matrigel, which was not needed in the migration assay. Afterwards, cells were incubated in serum-free 1640 medium for 24 h. Then, the cells were implanted in the upper chamber and the complete medium RPMI-1640 was loaded into the lower chamber. After 48-h incubation, the migrated and invaded cells were fixed with paraformaldehyde, stained with crystal violet and counted under a microscope (IX83, Olympus Corporation, Japan).
Dual-luciferase assay
The associations between LINC01410 and miR-23c, as well as miR-23c and CHD7 were identified by dual-luciferase assays. First, 3′-UTR of LINC01410 or 3′-UTR of CHD7 was inserted into pGL3 vector (Promega, Madison, WI) to construct the wild type (WT) luciferase reporter vector pGL3-LINC01410 or pGL3-CHD7. Afterwards, Site-Directed Mutagenesis Kit (210518, Stratagene, La Jolla, USA) was used to synthesis the mutated luciferase reporter vector pGL3-LINC01410-MUT or pGL3-CHD7-MUT with point mutations in the seed sequence. Then, luciferase vector and miR-23c mimic or mimic NC were co-transfected into cells. After 48 h of incubation, the cells were lysed and the luciferase activity in every group was evaluated using the dual-luciferase reporter assay system (Promega, Madison, WI).
Statistical analysis
All experiments in this study were acquired in triplicate independently. The values were exhibited as the means ± standard deviation (SD). Statistical analyses were assessed by using SPSS 22.0 and GraphPad Prism 5.0. The significant differences among groups were measured by student’s t test or one-way ANOVA accompanied by Bonferroni’s post hoc analysis. Low and high expression of LINC01410, miR-23c, and CHD7 in the EC samples were defined according to the median, and those above the median were defined as high expression and those under the median were defined as low expression. And the prognosis was analyzed by Kaplan–Meier Plotter. Clinical correlation analysis was performed using chi-square test. P value less than 0.05 revealed a statistically significance.
Results
LINC01410 expression was connected with the survival of EC patients and biological characteristics of EC cells
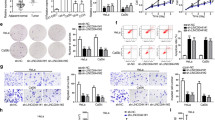
To search for the function of LINC01410 in EC development, we first assessed the expression of LINC01410 in EC tissues from TCGA database. The data indicated that LINC01410 expression was obviously increased in EC tissues (n = 552) in contrast with that of the control (n = 35, Fig. 1a, P < 0.0001), and high expression of LINC01410 resulted in lower survival of EC patients (Fig. 1b, P < 0.01). Moreover, LINC01410 expression was closely related to age, grade, histological type, stage and death of EC patients (Table 1). Afterwards, we detected the expression of LINC01410 in EC cell lines including HEC-1-A, RL95-2 and KLE. The data revealed that LINC01410 expression was elevated to varying degrees in EC cells compared to that of the normal cell NUEEC (Fig. 1c, P < 0.01). HEC-1-A and KLE cells were employed to perform loss or gain of function experiments. After si-LINC01410#1 or si-LINC01410#2 treatment, the expression of LINC01410 was reduced significantly in KLE cells (Fig. 1d, P < 0.01). In the subsequent loss of function experiments, si-LINC01410#2 with higher interference efficiency was selected. Besides, LINC01410 expression was obviously increased after pcDNA3.1-LINC01410 treatment in HEC-1-A cells (Fig. 1e, P < 0.01). And the data from cell proliferation assays exhibited that knockdown of LINC01410 decreased the viability of KLE cells, and upregulation of LINC01410 increased the viability of HEC-1-A cells (Fig. 1f, g, P < 0.01). Furthermore, the number of invaded and migrated KLE cells were significantly decreased after LINC01410 depletion, but overexpression of LINC01410 elevated the number of invaded and migrated HEC-1-A cells (Fig. 1h, i, P < 0.01). Besides, the expression of proteins associated with cell cycle and apoptosis was also detected by western blot. The data presented that depletion of LINC01410 reduced the expression of Cyclin D1, CDK6 and Bcl-2, and elevated the Bax expression, indicating that knockdown of LINC01410 blocked the cell cycle and promoted cell apoptosis (Fig. 1j, P < 0.01). All these phenomena suggested that LINC01410 possessed a crucial role in EC development.
High expression of LINC01410 was related to poor prognosis of EC patients and the growth, cycle, apoptosis, invasion and migration of EC cells. a LINC01410 was highly expressed in EC tissues (n = 552) compared with that of the normal tissues (n = 35). b EC patients with LINC01410 upregulation presented worse overall survival. c The LINC01410 expression was upregulated in EC cell lines. d LINC01410 expression was significantly reduced after si-LINC01410 treatment in KLE cells. e LINC01410 expression was obviously elevated after pcDNA3.1-LINC01410 transfection in HEC-1-A cells. f The viability of KLE cells was decreased after knockdown of LINC01410. g The viability of HEC-1-A was upregulated after LINC01410 overexpression. h Depletion of LINC01410 reduced the numbers of invaded and migrated KLE cells. i Upregulation of LINC01410 increased the number of invaded and migrated HEC-1-A cells. j Expression of cycle-related proteins and apoptosis-related proteins was detected after LINC01410 knockdown. P < 0.01 vs. control
Bioinformatics analysis predicted and constructed the ceRNA network related to LINC01410
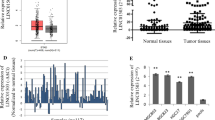
Next, we analyzed LINC01410 localization in cells through long non-coding RNA subcellular localization predictor (lnclocator) website, and found that LINC01410 was widely distributed in cytoplasm, which was speculated to be related to ceRNA mechanism (Fig. 2a). Then, lncBase website was applied to predict the miRNAs related to LINC01410, and 823 miRNAs were obtained. The predicted miRNAs were intermingled with the downregulated miRNAs with prognostic significance, and 6 miRNAs were captured, namely miR-23c, miR-4424, miR-47045, miR-4770, miR-548ax, and miR-578 (Fig. 2b). Based on the literature review, miR-23c was further analyzed as a downstream miRNA of LINC01410. The information from TCGA revealed that miR-23c was lower expressed in EC tissues (n = 546) compared with that of the normal tissues (n = 33), and its underexpression easily led to unfavorable prognosis (Fig. 2c, d, P < 0.01). In addition, miR-23c expression was associated with the age, grade, histological type, stage and death of EC patients (Table 2). Afterwards, the downstream target of miR-23c was investigated. TargetScan website was used to predict the target genes of miR-23c, and 1343 genes were acquired. The predicted target genes were intersected with mRNAs that were upregulated and had significant prognostic values, and 24 genes were obtained (Fig. 2e). The 24 target genes of miR-23c were intersected with the LINC01410 co-expressed genes, and 5 genes were acquired, namely SOX11, LIN28B, EPHB2, STON2 and CHD7 (Fig. 2f). Through the literature review, CHD7 was selected for further research, which has relatively few literature studies. The data from TCGA suggested that CHD7 was highly expressed in EC tissues (n = 552) compared with that of the normal tissues (n = 35), and high expression of CHD7 caused worse overall survival in patients with EC (Fig. 2g, h). And CHD7 expression was connected with the age, grade, histological type, and death of EC patients (Table 3). Considering the relationship among LINC01410, miR-23c and CHD7, the effects of LINC01410/miR-23c/CHD7 combination on survival were performed. The data showed that patients with high-LINC01410/CHD7 expression and low-miR-23c expression had worse prognosis than other patients (Fig. 2i). However, patients with low-LINC01410/CHD7 expression and high-miR-23c expression had better prognosis than other patients (Fig. 2j). These results insinuated that LINC01410/miR-23c/CHD7 may form a ceRNA network to affect the biological behaviors of EC samples.
Associations among LINC01410, miR-23c and CHD7 were predicted by bioinformatics analysis. a The lnclocator website was used to detect the location of LINC01410 in cells. b MiRNAs, related to LINC01410, were obtained. c The expression of miR-23c in EC tissues (n = 546) were presented by analysis of TCGA database. d The overall survival of EC patients with miR-23c downregulation became shorter. e, f The mRNAs, related to miR-23c, were acquired. g The expression of CHD7 was significantly upregulated in GC tissues (n = 552) compared with that of the normal tissues (n = 35). h Patients with CHD7 upregulation presented poor overall survival. i Patients with high-LINC01410/CHD7 expression and low miR-23c expression exhibited worse prognosis than other patients. j Patients with low-LINC01410/CHD7 expression and high miR-23c expression showed better prognosis than other patients. P < 0.01 vs. normal
LINC01410 promoted CHD expression, at least in part, by regulating miR-23c expression in EC cells
To further identify the associations among LINC01410, miR-23c and CHD7, dual-luciferase assays were carried out. The binding sites between LINC01410 and miR-23c, as well as miR-23c and CHD7 were presented in Fig. 3a, b. The luciferase activity in WT LINC01410 group was decreased or increased after treatment with miR-23c mimic or inhibitor, but the activity in MUT LINC01410 group was almost the same (Fig. 3c, P < 0.01). And the luciferase activity in WT CHD7 group was reduced or elevated after up or downregulation of miR-23c, but the activity in MUT CHD7 group was almost not change (Fig. 3d, P < 0.01). And the data from PCR and western blot exhibited that CHD7 expression was decreased or increased after LINC01410 or miR-23c depletion, but knockdown of LINC01410 and miR-23c together attenuated the effects of si-LINC01410 or miR-23c inhibitor on CHD7 expression (Fig. 3e–g, P < 0.01). Moreover, upregulation of LINC01410 or miR-23c elevated or reduced the expression of CHD7, but simultaneous overexpression of LINC01410 and miR-23c suppressed the effects of LINC01410-OE or miR-23c mimic on CHD7 expression (Fig. 3h–j, P < 0.01). These data illustrated that CHD7 expression was positively modulated by LINC01410, and inversely regulated by miR-23c, affording basis for further exploration.
Associations among LINC01410, miR-23c and CHD7 were identified by biological experiments. a, b The sequences of WT 3′ UTR of LINC01410, MUT 3′ UTR of LINC01410, miR-23c, WT 3′ UTR of CHD7, MUT 3′ UTR of CHD7 were presented. c The luciferase activity in WT LINC01410 was altered after miR-23c mimic or inhibitor treatment, **P < 0.01 vs. NC. d The luciferase activity in WT CHD7 was altered after miR-23c mimic or inhibitor treatment, **P < 0.01 vs. control. e–g. The mRNA and protein expression of CHD7 in KLE cells were assessed after different treatment, **P < 0.01 vs. control, ##P < 0.01 vs. si-LINC01410+ inhibitor. h–j The mRNA and protein expression of CHD7 in HEC-1-A cells were assessed after different treatment, **P < 0.01 vs. control, ##P < 0.01 vs. LINC01410-OE+ mimic
LINC01410 regulated the activity, invasion and migration of EC cells via interacting with miR-23c/CHD7
Based on the above research, the regulation of LINC01410/miR-23c/CHD7 axis on the activity, invasion and migration of EC cells was further detected. We observed that the activity of KLE cells was increased significantly after miR-23c depletion or CHD7 upregulation. But depletion of LINC01410 suppressed the promoting effects of miR-23c inhibitor or CHD7-OE on cells viability. Together overexpression of CHD7 and silencing of miR-23c enhanced the promoting effects of miR-23c inhibitor or CHD7-OE on KLE cells viability (Fig. 4a, P < 0.01). On the other side, overexpression of miR-23c or depletion of CHD7 reduced the viability of HEC-1-A cells. However, upregulation of LINC01410 limited the inhibitory effects of miR-23c mimic or si-CHD7 on cell viability. Moreover, simultaneous upregulation of miR-23c and silencing of CHD7 strengthened the inhibitory effects of miR-23c mimic or si-CHD7 on HEC-1-A cells vitality (Fig. 4b, P < 0.01). The transwell assays stated that knockdown of miR-23c or upregulation of CHD7 increased the numbers of invaded and migrated KLE cells. But knockdown of LINC01410 limited the promoting effects of miR-23c inhibitor or CHD7-OE on invasion and migration of KLE cells. Upregulation of CHD7 and depletion of miR-23c strengthened the promoting effects of miR-23c inhibitor or CHD7-OE on KLE cells invasion and migration (Fig. 4c, P < 0.01). Whilst, overexpression of miR-23c or depletion of CHD7 decreased the invasion and migration numbers of HEC-1-A cells. However, overexpression of LINC01410 suppressed the inhibitory effects of miR-23c mimic or si-CHD7 on HEC-1-A cells invasion and migration. In addition, upregulation of miR-23c and knockdown of CHD7 enhanced the inhibitory effects of miR-23c mimic or si-CHD7 on HEC-1-A cells invasion and migration (Fig. 4d, P < 0.01). These data indicated that LINC01410/miR-23c/CHD7 formed a ceRNA network to regulate the proliferation, invasion and migration of EC cells.
LINC01410, miR-23c and CHD7 formed a ceRNA network to regulate EC cells biological characteristics. a The activity of KLE cells was assessed by CCK-8 assay after different treatment, **P < 0.01 vs. control, ##P < 0.01 vs. miR-23c inhibitor, &&P < 0.01 vs. CHD7-OE. b The activity of HEC-1-A cells was evaluated by CCK-8 assay after different treatment, **P < 0.01 vs. control, ##P < 0.01 vs. miR-23c mimic, &&P < 0.01 vs. si-CHD7. c The invasion and migration of KLE cells were changed after different treatment, **P < 0.01 vs. control, ##P < 0.01 vs. miR-23c inhibitor, &&P < 0.01 vs. CHD7-OE. d The invasion and migration of HEC-1-A cells were changed after different treatment, **P < 0.01 vs. control, ##P < 0.01 vs. miR-23c mimic, &&P < 0.01 vs. si-CHD7
Discussion
The crucial roles of non-coding RNAs have become a focus of study on EC tumorigenic mechanisms. Non-coding RNAs, as upstream regulators of protein function, structure, and signaling pathways related to human disease and organ development, were also widely regarded as important targets for EC therapy [23]. Numerous lncRNAs have been identified as oncogenes or tumor-suppressive genes involved in the development of EC. Interestingly, we observed that LINC01410 was highly expressed in EC tissues, and related to the worse prognosis of EC patients. And clinical correlation analysis exhibited that LINC01410 expression was correlated with age, histological grade, pathological type, clinical stage and death of EC patients. In addition, we used LINC01410 to construct a ceRNA network, and discovered that LINC01410 promoted the malignant behaviors of EC cells by regulating miR-23c/CHD7 axis.
In cancer research, lncRNAs and miRNAs are the most common non-coding RNAs, which are important components of tumor biology [24]. It has been appreciated that lncRNAs conduce to the development and progression of numerous types of tumors [25], including EC. For example, the following lncRNAs including LOC134466, OGFRP1, PVT1, and NEAT1 have been discovered to be upregulated in EC and promoted the progression of EC [4, 20, 26, 27]. By exploring TCGA database, LINC01410 was selected for further research due to its high expression in EC. LINC01410 was reported to possess crucial roles in numerous biological behaviors, covering cell cycle, apoptosis, propagation, migration. For example, LINC01410 depletion suppressed the proliferation and migration of colon cancer cells, and blocked the cells in G0/G1 phase [16]. In addition, LINC01410 silencing blocked cells growth and promoted apoptosis in papillary thyroid carcinoma [28]. Importantly, our present study found that LINC01410, which was highly expressed in EC samples, may act as an oncogene in EC through promoting cell cycle, growth and invasion, as well as suppressing cell apoptosis, and that suppression of LINC01410 might hold a post of the promising therapeutic approach in EC treatment. To discover the potential pro-oncogenic mechanism of LINC01410, we identified miR-23c as a novel target of LINC01410. MiR-23c, emerged as a cancer-associated miRNA, has been demonstrated to suppress tumor growth of hepatocellular carcinoma [22]. Moreover, miR-23c, as a downstream target of hsa_circ_0018069, played crucial roles in bladder cancer [29]. Besides, the presence of low rate of miR-23c/DMBX1 in EC may have a better prediction value through bioinformatics analysis from Xu et al. [30]. Given this, we speculated that miR-23c may play an important role in EC development. Through analysis, miR-23c was demonstrated to be lower expressed in EC tissues, which was associated with poor prognosis and clinical correlation. Furthermore, the inhibitory effects of miR-23c mimic on EC cells growth and aggressiveness were rescued by LINC01410 upregulation.
As bioformatics analysis and luciferase assays suggested, CHD7 was verified as a target of miR-23c that influenced EC malignancy. CHD7, as an ATP-dependent chromatin remodeler, is necessary for neural differentiation based on its transcriptional modulation in progenitor cells [31]. Besides, several studies revealed that CHD7 deficiency delays leukemogenesis in mice caused by Cbfb-MYH11 [32], and CHD7, which higher expression was associated with poor prognosis of patients in pancreatic cancer, was also dysregulated in some cases of pancreatic cancer [33]. In medulloblastoma, CHD7 lower expression is a signature of patients with worse overall survival [34]. In line with previous study in pancreatic cancer, our research discovered that CHD7 was highly expressed in EC tissues, and its high expression was related to poor prognosis of EC patients and clinical significance. CHD7 expression was regulated by miR-23c and LINC01410. And the promoting effects of CHD7 on EC cells growth and aggressiveness were strengthened by LINC01410 and limited by miR-23c. These findings insinuated that LINC01410, miR-23c and CHD7 formed a ceRNA network to regulate the growth and aggressiveness of EC cells. Although some results were acquired from the research, the shortcoming of the study should be clarified. First, the results lack evidence for in vivo experiments. Second, not enough markers of malignant behaviors were detected.
In summary, our research discovered that LINC01410 functioned as a positive regulator in EC progression through sponging miR-23c to upregulate CHD7, providing novel molecules for EC-targeted therapy and prognosis. Based on our results, we proposed a new molecular mechanism of LINC01410 in regulation of EC cells growth and invasion (Fig. 5).
References
McAlpine JN, Temkin SM, Mackay HJ (2016) Endometrial cancer: not your grandmother's cancer. Cancer 122:2787–2798
Temkin SM, Minasian L, Noone AM (2016) The end of the hysterectomy epidemic and endometrial cancer incidence: what are the unintended consequences of declining hysterectomy rates? Front Oncol 6:89
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68:394–424
Xu H, Sun Y, Ma Z, Xu X, Qin L, Luo B (2018) LOC134466 methylation promotes oncogenesis of endometrial carcinoma through LOC134466/hsa-miR-196a-5p/TAC1 axis. Aging (Albany NY) 10:3353–3370
Tsai KW, Lo YH, Liu H, Yeh CY, Chen YZ, Hsu CW, Chen WS, Wang JH (2018) Linc00659, a long noncoding RNA, acts as novel oncogene in regulating cancer cell growth in colorectal cancer. Mol Cancer 17:72
Ye B, Liu B, Yang L, Zhu X, Zhang D, Wu W, Zhu P, Wang Y, Wang S, Xia P, Du Y, Meng S, Huang G, Wu J, Chen R, Tian Y, Fan Z (2018) LncKdm2b controls self-renewal of embryonic stem cells via activating expression of transcription factor Zbtb3. EMBO J. https://doi.org/10.15252/embj.201797174
Wu X, He X, Li S, Xu X, Chen X, Zhu H (2016) Long non-coding RNA ucoo2kmd.1 regulates CD44-dependent cell growth by competing for miR-211-3p in colorectal cancer. PLoS ONE 11:e0151287
Spizzo R, Almeida MI, Colombatti A, Calin GA (2012) Long non-coding RNAs and cancer: a new frontier of translational research? Oncogene 31:4577–4587
Ma H, Hao Y, Dong X, Gong Q, Chen J, Zhang J, Tian W (2012) Molecular mechanisms and function prediction of long noncoding RNA. Sci World J 2012:541786
Sun MY, Zhu JY, Zhang CY, Zhang M, Song YN, Rahman K, Zhang LJ, Zhang H (2017) Autophagy regulated by lncRNA HOTAIR contributes to the cisplatin-induced resistance in endometrial cancer cells. Biotechnol Lett 39:1477–1484
Xie P, Cao H, Li Y, Wang J, Cui Z (2017) Knockdown of lncRNA CCAT2 inhibits endometrial cancer cells growth and metastasis via sponging miR-216b. Cancer Biomark 21:123–133
Chen S, Wang LL, Sun KX, Liu Y, Guan X, Zong ZH, Zhao Y (2018) LncRNA TDRG1 enhances tumorigenicity in endometrial carcinoma by binding and targeting VEGF-A protein. Biochim Biophys Acta Mol Basis Dis 1864:3013–3021
Zhang G, Ma A, Jin Y, Pan G, Wang C (2019) LncRNA SNHG16 induced by TFAP2A modulates glycolysis and proliferation of endometrial carcinoma through miR-490-3p/HK2 axis. Am J Transl Res 11:7137–7145
Sun KX, Wu DD, Chen S, Zhao Y, Zong ZH (2017) LncRNA MEG3 inhibit endometrial carcinoma tumorigenesis and progression through PI3K pathway. Apoptosis 22:1543–1552
Zhang JX, Chen ZH, Chen DL, Tian XP, Wang CY, Zhou ZW, Gao Y, Xu Y, Chen C, Zheng ZS, Weng HW, Ye S, Kuang M, Xie D, Peng S (2018) LINC01410-miR-532-NCF2-NF-kB feedback loop promotes gastric cancer angiogenesis and metastasis. Oncogene 37:2660–2675
Luo J, Guo Y, Liu X, Yang X, Xiao F, Zhou M (2018) Long non-coding RNA LINC01410 promotes colon cancer cell proliferation and invasion by inhibiting miR-3128. Exp Ther Med 16:4824–4830
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP (2011) A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146:353–358
Li JH, Liu S, Zhou H, Qu LH, Yang JH (2014) starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 42:D92–D97
Yang S, Ning Q, Zhang G, Sun H, Wang Z, Li Y (2016) Construction of differential mRNA-lncRNA crosstalk networks based on ceRNA hypothesis uncover key roles of lncRNAs implicated in esophageal squamous cell carcinoma. Oncotarget 7:85728–85740
Lv Y, Chen S, Wu J, Lin R, Zhou L, Chen G, Chen H, Ke Y (2019) Upregulation of long non-coding RNA OGFRP1 facilitates endometrial cancer by regulating miR-124-3p/SIRT1 axis and by activating PI3K/AKT/GSK-3beta pathway. Artif Cells Nanomed Biotechnol 47:2083–2090
Muhammad N, Bhattacharya S, Steele R, Ray RB (2016) Anti-miR-203 suppresses ER-positive breast cancer growth and stemness by targeting SOCS3. Oncotarget 7:58595–58605
Zhang L, Wang Y, Wang L, Yin G, Li W, Xian Y, Yang W, Liu Q (2018) miR-23c suppresses tumor growth of human hepatocellular carcinoma by attenuating ERBB2IP. Biomed Pharmacother 107:424–432
Zhou YX, Wang C, Mao LW, Wang YL, Xia LQ, Zhao W, Shen J, Chen J (2018) Long noncoding RNA HOTAIR mediates the estrogen-induced metastasis of endometrial cancer cells via the miR-646/NPM1 axis. Am J Physiol Cell Physiol 314:C690–c701
Huang YK, Yu JC (2015) Circulating microRNAs and long non-coding RNAs in gastric cancer diagnosis: An update and review. World J Gastroenterol 21:9863–9886
Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, Wang Y, Brzoska P, Kong B, Li R, West RB, van de Vijver MJ, Sukumar S, Chang HY (2010) Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464:1071–1076
Li Z, Wei D, Yang C, Sun H, Lu T, Kuang D (2016) Overexpression of long noncoding RNA, NEAT1 promotes cell proliferation, invasion and migration in endometrial endometrioid adenocarcinoma. Biomed Pharmacother 84:244–251
Kong F, Ma J, Yang H, Yang D, Wang C, Ma X (2018) Long non-coding RNA PVT1 promotes malignancy in human endometrial carcinoma cells through negative regulation of miR-195-5p. Biochim Biophys Acta Mol Cell Res 1865:1479–1490
Wang G, Wang X, Jin Y (2019) LINC01410/miR-3619-5p/FOXM1 feedback loop regulates papillary thyroid carcinoma cell proliferation and apoptosis. Cancer Biother Radiopharm 34:572–580
Li M, Wang Y, Liu Y, Zhang X, Liu J, Wang P (2019) Low expression of hsa_circ_0018069 in human bladder cancer and its clinical significance. Biomed Res Int 2019:9681863
Xu X, Liu T, Wang Y, Fu J, Yang Q, Wu J, Zhou H (2019) miRNA-mRNA associated with survival in endometrial cancer. Front Genet 10:743
Feng W, Shao C, Liu HK (2017) Versatile roles of the chromatin remodeler CHD7 during brain development and disease. Front Mol Neurosci 10:309
Zhen T, Kwon EM, Zhao L, Hsu J, Hyde RK, Lu Y, Alemu L, Speck NA, Liu PP (2017) Chd7 deficiency delays leukemogenesis in mice induced by Cbfb-MYH11. Blood 130:2431–2442
Colbert LE, Petrova AV, Fisher SB, Pantazides BG, Madden MZ, Hardy CW, Warren MD, Pan Y, Nagaraju GP, Liu EA, Saka B, Hall WA, Shelton JW, Gandhi K, Pauly R, Kowalski J, Kooby DA, El-Rayes BF, Staley CA 3rd, Adsay NV, Curran WJ Jr, Landry JC, Maithel SK, Yu DS (2014) CHD7 expression predicts survival outcomes in patients with resected pancreatic cancer. Cancer Res 74:2677–2687
Badodi S, Dubuc A, Zhang X, Rosser G, Da Cunha JM, Kameda-Smith MM, Morrissy AS, Guilhamon P, Suetterlin P, Li XN, Guglielmi L, Merve A, Farooq H, Lupien M, Singh SK, Basson MA, Taylor MD, Marino S (2017) Convergence of BMI1 and CHD7 on ERK signaling in medulloblastoma. Cell Rep 21:2772–2784
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lu, M., Ding, N., Zhuang, S. et al. LINC01410/miR-23c/CHD7 functions as a ceRNA network to affect the prognosis of patients with endometrial cancer and strengthen the malignant properties of endometrial cancer cells. Mol Cell Biochem 469, 9–19 (2020). https://doi.org/10.1007/s11010-020-03723-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-020-03723-9