Abstract
Long noncoding ribonucleic acids (lncRNAs) are critical regulators in various biological processes. In the present study, we aimed to explore whether miR140-3p was involved in the underlying molecular mechanisms of small nucleolar RNA host gene 1 (SNHG1) in myocardial ischemia/reperfusion (I/R) injury. A mouse model of I/R injury and hypoxia-reoxygenation (H/R)-stimulated human umbilical vein endothelial cells (HUVECs) was used in this study. Cell proliferation was detected by MTT. The mRNA and protein levels of vascular endothelial growth factor (VEGF), VE-cadherin, and MMP2 were detected by RT-PCR and western blot, respectively. The angiogenesis was assessed by tube formation assay. Cell migration was assessed using wound-healing assay. Results showed that SNHG1 expression was increased in the cardiac microvasculature of a mouse model of I/R injury and in H/R-stimulated HUVECs. H/R stimulation significantly reduced cell proliferation, tube formation, and cell migration, but increased expression of VEGF, VE-cadherin, and MMP2. SNHG1 upregulation under H/R increased HUVECs proliferation, tube formation, and cell migration, and upregulated expression of VEGF, VE-cadherin, and MMP2, compared with the H/R group. SNHG1 knockdown exhibited the opposite effect. SNHG1 functioned as a competing endogenous RNA (ceRNA) of miR-140-3p. HIF-1α was identified as a target of miR-140-3p. SNHG1 upregulation enhanced cell proliferation, tube formation, and expression of VEGF, VE-cadherin, and MMP2 through HIF-1α/VEGF signaling. This process could be offset by miR-140-3p mimic or VEGF inhibitor. Our results reveal a novel protective function of SNHG1 that furthers understanding of cardiac I/R injury and provides experimental evidence for future therapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Myocardial ischemia/reperfusion (I/R) injury can result in severe myocardial damage and contributes to high mortality [1]. Researchers have dedicated to reducing various pathophysiological components of I/R injury to limit cell death. However, it is still unclear how gene expression is regulated during I/R.
LncRNAs function in various biological processes. They regulate gene expression through several mechanisms, such as modulating transcription factor activity, co-activating with neighboring genes, and acting as transcription enhancers [2, 3]. RNA-seq analyses showed that ischemia dramatically alters series of lncRNAs expression in ischemic-induced cerebrovascular dysfunction and myocardial injury [3, 4]. LncRNA H19 has been reported to regulate endothelial cell aging via inhibition of Stat3 signaling [5], lncRNA malat1 regulates angiogenesis in hindlimb ischemia [6], and lncRNA small nucleolar RNA host gene 16 (SNHG16) drives proliferation, migration, and invasion of hemangioma endothelial cells by modulating the miR-520d-3p/STAT3 axis [7].
LncRNA SNHG1 (gene ID: 23642) is localized at 11q12.3 and has 11 exons. SNHG1 expression was upregulated in gastric, colorectal, liver, and lung cancer [8]. Not only could this lncRNA regulate SLC3A2 expression through binding with mediator complex, but also it directly targeted FUBP1 to promote cancer cell proliferation [8]. In brain microvascular endothelia, SNHG1 expression was induced by oxygen–glucose deprivation [4, 9]. SNHG1 overexpression alleviated cardiomyocyte injury induced by hydrogen peroxide via regulating miR-195/BCL2L2 interaction [10]. Therefore, we are wondering whether it also plays a role in myocardial I/R injury.
I/R caused microvascular injury, which is associated with high morbidity and mortality. Altered wall thickness, damaged endothelial nuclei, and reduced cell junction density were observed in coronary microcirculation after I/R surgery [11]. Leukocyte adhesion to vascular endothelial and inflammation after myocardial infarction would result in left-ventricular dysfunction [12]. Vascular endothelial dysfunction after I/R was induced through many factors including pH alteration, oxidative stress, nitric oxide inhibition, ion channels disruption, and gap junction destruction [13]. In the present study, with a mouse model of I/R injury and hypoxia-reoxygenation-stimulated human umbilical vein endothelial cells (HUVECs), we would examine the expression and the role of SNHG1 in cardiac endothelial injury induced by I/R or hypoxia/reoxygenation (H/R).
Materials and methods
Antibodies and reagents
All substances were purchased from Gibco (Grand Island, NY, USA) unless otherwise stated. Rabbit anti-MMP2 antibodies were purchased from Abcam (Cambridge, MA, USA). Antibodies against vascular endothelial growth factor (VEGF) were bought from Sigma (St. Louis, MO, USA). The monoclonal antibody against vascular endothelial cadherin (VE-cadherin) was obtained from Cell Signaling Technology (Beverly, MA, USA). Horseradish-peroxidase-conjugated goat anti-mouse and anti-rabbit antibodies were from Cell Signaling Technology (San Diego, CA, USA).
Animals
All experimental protocols were reviewed and approved by the Fourth Military Medical University committee. The study was conducted in accordance with the Declaration of Helsinki. C57BL/6 female mice aged 8–10 weeks were purchased from the Experimental Animal Center of Peking University, Beijing (license SCXK [Jing] 2011-0012), and were housed under controlled temperature (24 ± 2 °C) and subjected to a natural photoperiod (12 h light/dark cycle). They were allowed free access to food and water throughout the experiment period.
Myocardial ischemia/reperfusion injury model
Briefly, mice were pre-medicated with heparin (1000 IU/kg, i.p.) and anesthetized 5 min later with sodium pentobarbital (60 mg/kg, i.p.). Following the left thoracotomy between the fourth and fifth ribs, the left anterior descending coronary artery was visualized under a microscope and ligated using a 6.0 Prolene suture. Regional ischemia was confirmed by visual inspection under a dissecting microscope (Leica) of discoloration of the occluded distal myocardium. For I/R, the ligation was released after 45 min of ischemia. Sham-operated animals underwent the same procedure without occlusion of the left anterior descending coronary artery [14].
Cell culture and treatment
HUVECs were purchased from ScienCell (San Diego, CA, USA) and cultured in cardiac myocyte medium supplemented with 5% fetal bovine serum and 1% penicillin/streptomycin at 37 °C in a humidified incubator with 95% atmosphere and 5% CO2 according to the manufacturer’s instructions. We used HUVECs at passages 7–10 for the experiments. After serum starvation, cells were transferred into a hypoxic incubator containing 1% O2, 5% CO2, and 94% N2 with ischemia-mimetic solution. Cells were subjected to hypoxic condition for 2 h and then reoxygenated by incubating in fresh cardiac myocyte medium and rapidly transferring to a normoxic incubator (20% O2, 5% CO2, and 75% N2) for 4 h.
Plasmid constructs and transfection
SNHG1 cDNA was amplified and subcloned in the Kpn I and BamH I sites of pcDNA3.1 (Invitrogen, Carlsbad, CA, USA) to construct the pcDNA3.1-SNHG1 vector. The specific primer of SNHG1 was synthesized by Sangon Biotech (Shanghai, China). To construct the si-SNHG vector, the self-complementary hairpin DNA oligonucleotides were annealed and ligated into the pSUPER plasmid vector. Negative control was named as the mock vector. The miR-140-3p mimic was used to overexpress miR-140-3p, and its control was named mimic-nc. Anti-miR-140-3p was used to downregulate miR-140-3p expression, with the anti-control (anti-nc) as a control. The si-SNHG, mock, miR-140-3p mimic, mimic-nc, anti-miR-140-3p, or anti-nc were transfected into HUVECs using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA).
Cell proliferation assay
Cell proliferation was determined using the conventional MTT assay. Cells grown in 96-well microplates at a density of 1 × 104 cells per well were exposed to H/R and the miR-410 mimic or miR-410 inhibitor. Then, the MTT solution (Sigma) was added to the culture medium to reach a final concentration of 0.5 mg/ml for 4 h at 37 °C. Absorbance measurements were read at a wavelength of 450 nm. The optical density value was reported as the percentage of the control group (set as 100%).
ELISA analysis
Serum levels of creatine kinase-MB (CK-MB) and lactate dehydrogenase (LDH) were analyzed using ELISA kits (Yanjin Biotechnology Co., Shanghai, China) according to the manufacturer’s procedure. All samples were analyzed in duplicate.
Tube formation assay
A 24-well culture plate was coated with 100 μl of 10 mg/ml prechilled Matrigel (Corning, Bedford, MA) plus VEGF (100 ng/ml). The HUVECs were seeded on the Matrigel-coated plate at concentrations of 5 × 104 cells per well. After 24-h culturing, images were acquired using an optical microscope (×100 magnification). The average length of the tube branches in four areas of each sample was assessed under an inverted light microscope.
Wound-healing assay
HUVECs cultured in 6-well culture plates were scratched using a 10-μl pipette tip when the cell density in each well reached 80–90% confluence. The wounded monolayers were washed with PBS to remove detached cells. Cells were then cultured at 37 °C for wound closure. Images were acquired at 0 h and 48 h after scratching using an optical microscope (×100 magnification). The percentage of the wound closure area in five randomly selected fields was calculated under an inverted light microscope.
Western blot
Proteins were extracted using a radioimmunoprecipitation assay (RIPA) buffer and then separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene fluoride membranes (Millipore, Billerica, MA, USA). The membranes were blocked with 5% fat-free milk powder and then exposed to antibodies against HIF-1α (1:1000), VEGF (1:1500) or β-actin (1:2000). The horseradish-peroxidase-conjugated corresponding secondary antibody was used at 1:5000. The enhanced chemiluminescence (Amersham, Little Chalfont, UK) was used for detection. Densitometry values were normalized to levels of β-actin and the control lane in each blot.
Quantitative real-time polymerase chain reaction (qRT-PCR) for miRNA and mRNA expression
Total RNA from HUVECs was extracted using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. The PCR primers specific for HIF-1α were 5′-CCC AAT GTC GGA GTT TGG AA-3′ and 5′-TGG CTG CAT CTC GAG ACT TT-3′; VEGF, 5′-GCT CAG AGC GGA GAA AGC AT-3′ and 5′-GCC TCG GCT TGT CAC ATC T-3′; SNHG1, 5′-CTG CTC GTG ATG TTC AGC C-3′ and 5′-AAT ACC TGT ATT CAC CCT GGG A-3′ (human); 5′-TCC TTG TTC GGG GTT TGA GG-3′ and 5′-ACA GCA CCC TGA CTA CAA GC-3′ (mouse); miR-140-3p, 5′-ACA CTC CAG CTG GGC AGT GGT TTT ACC CTA-3′ and 5′-TGG TGT CGT GGA GTC G-3′; U6 were 5′-CTC GCT TCG GCA GCA CA-3′ and 5′-AAC GCT TCA CGA ATT TGC GT-3′; β-actin, 5′-CTT CCA GCC TTC CTT CCT GG-3′ and 5′-TTC TGC ATC CTG TCG GCA AT-3′ (human); 5′-CTC CTA TGT GGG TGA CGA GG-3′ and 5′-ACG GTT GGC CTT AGG GTT C-3′ (mouse). Data were analyzed using the formula R = 2−[ta sample−ΔCt control]. β-Actin and U6 SnRNA were used as internal controls to form RNA and miRNA, respectively. PCRs were conducted in duplicate.
Luciferase assay
A truncated fragment of the SNHG1/HIF-1α 3′-untranslated region (UTR) containing the wild-type miR-140-3p predicted binding site (SNHG1/HIF-1α-WT) was cloned into the 3′ UTR of the firefly luciferase structural gene in the pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega, Madison, WI, USA). We also constructed separate plasmids containing the SNHG1/HIF-1α 3′ UTR with a mutated seed region for the predicted miR-140-3p binding site (SNHG1/HIF-1α-Mut) as the negative control. At 24 h before transfection, HUVECs cells were seeded on a 24-well plate (8 × 104 cells/well). Cells were co-transfected with 0.3 μg of the respective plasmid and 40 nM of the miR-140-3p mimic or mimic-nc using Lipofectamine 2000 (Invitrogen). At 48 h after plasmid vector transfection, the luciferase reporter assay was performed using a dual-luciferase assay kit (Promega) according to the manufacturer’s protocol. Data are presented as the ratio of firefly to Renilla luciferase activity.
RNA immunoprecipitation assay
RNA immunoprecipitation was implemented using an EZ-Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Billerica, MA, USA). Cells were lysed into complete RNA immunoprecipitation lysis buffer. Then, 100 μl of cell lysate was incubated with RNA immunoprecipitation buffer containing magnetic beads conjugated with human anti-Ago2 antibody (1:50 dilution) and negative control normal mouse IgG. Samples were incubated with proteinase K buffer, and then the target RNA was extracted for further study.
Data analysis
All the data were presented as mean ± SD and analyzed by using SPSS 20.0 software (SPSS Inc., Chicago, IL, USA). Student’s unpaired t tests or one-way ANOVA were performed to analyze the difference between two groups or more than two groups. Differences were considered statistically significant at P < 0.05.
Results
Altered SNHG1 levels in vivo after cardiac I/R injury
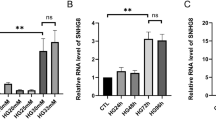
Serum levels of CK-MB and LDH were indicators for animals with cardiac I/R injury. We found that increased secretion of CK-MB and LDH was presented in mouse serum at 24 h after I/R (Fig. 1a). In the following, SNHG1 expression in mouse cardiac endothelial cells was analyzed at 6–72 h after I/R. The results showed that SNHG1 level was enhanced at 6 h and peaked at 12 h after I/R (Fig. 1b). Interestingly, SNHG1 levels were then significantly decreased at 24, 48, and 72 h after I/R (Fig. 1b).
SNHG1 levels in vivo after cardiac I/R injury was increased. a Serum levels of CK-MB and LDH in mouse cardiac endothelial cells were assessed by ELISA kits at 24 h after I/R (n = 6). b SNHG1 levels in mouse cardiac endothelial cells were assessed by RT-PCR at 6 h, 12 h, 24 h, 48 h, 72 h after I/R, respectively (n = 6). The data were presented as mean ± SD. *P < 0.05, compared with sham group. #P < 0.05, compared with 12 h after H/R
SNHG1 modulated proliferation and function of HUVECs
Furthermore, we explored SNHG1 level in cultured HUVECs, which were exposed to an H/R condition. H/R exposure slightly reduced SNHG1 level in these cells (Fig. 2a). To further investigate the role of SNHG1 in HUVECs, cells were transfected with SNHG1 overexpression vector or siRNA vector (Fig. 2a). MTT analysis of the cultured HUVECs showed that, compared with the control group, cell proliferation was significantly decreased by H/R, but was partly offset by SNHG1 upregulation (Fig. 2b). SNHG1 knockdown further decreased cell proliferation (Fig. 2b). H/R significantly increased expression of VEGF, VE-cadherin, and MMP2 (Fig. 2c–e). SNHG1 overexpression promoted the changes induced by H/R, whereas SNHG1 knockdown significantly reduced the expression of VEGF, VE-cadherin, and MMP2 (Fig. 2c–e). Results of the tube formation assays showed that H/R significantly decreased total tube length and that SNHG1 overexpression effectively promoted total tube length in vitro, whereas SNHG1 knockdown significantly decreased total tube length (Fig. 2f, g). In the wound-healing assay, H/R significantly decreased wound closure (Fig. 2h, i). SNHG1 overexpression significantly increased, but SNHG1 knockdown significantly decreased wound closure (Fig. 2h, i). These results suggest an association between SNHG1 and angiogenesis and cell migration.
SNHG1-modulated proliferation and function of HUVECs. a HUVECs were transfected with SNHG1, si-SNHG1, or corresponding controls (blank or mock). The transfection efficiency was detected by RT-PCR. b Cell proliferation was determined by MTT assay. The mRNA and protein levels of vascular endothelial growth factor, VE-cadherin, and MMP2 were determined by RT-PCR (c) and western blot (d), respectively. e The quantitative data of western blot. f The representative charts of tube formation in tube formation assay. g The quantitative data of tube formation. h The representative images of wound-healing assay. i The quantitative data of the percentage of the wound closure area. The data were presented as mean ± SD. *P < 0.05, compared with corresponding negative controls. #P < 0.05, compared with H/R group
MiR-140-3p was a target of SNHG1
Previous study has shown that miR-140-3p played a role in ventricular remodeling 28922712. Here, we found that miR-140-3p expression in cardiac endothelial cells was suppressed at 12 h after I/R (Fig. 3a). However, its expression was then increased after 24 h (Fig. 3a).
MiR-140-3p was a targeted target of SNHG1. a RT-PCR analysis of miR-140-3p expression in the cardiac microvasculature of a mouse model of cardiac I/R injury (n = 6). *P < 0.05, compared with sham group. #P < 0.05, compared with 12 h after H/R. b The putative miR-140-3p binding sequence of the wild-type and mutation sequence of SNHG1. c Relative luciferase assays. *P < 0.05, compared with mimic-nc group. d RT-PCR analysis of miR-140-3p expression in HUVECs that were transfected with SNHG1, si-SNHG1, or negative controls (blank or mock). *P < 0.05, compared with the control group. #P < 0.05, compared with the H/R group. The data were presented as mean ± SD
Bioinformatics analyses showed that miR-140-3p could bind with one conserved site of SNHG1 (Fig. 3b). Moreover, the result of luciferase reporter assay showed that miR-140-3p mimic reduced the luciferase activity of pGL3-SNHG1 but not pGL3-SNHG1-MUTANT (Fig. 3c). Additionally, SNHG1 overexpression downregulated, while SNHG1 knockdown upregulated miR-140-3p expression in HUVECs, suggesting that SNHG1 could regulate miR-140-3p expression through the interaction with miR-140-3p (Fig. 3d).
SNHG1 modulated proliferation and function of HUVECs by negatively regulating miR-140-3p
To investigate the role of miR-140-3p in hypoxia-induced vascular endothelial cell injury, miR-140-3p expression was upregulated or downregulated in HUVECs (Fig. 4a). SNHG1 overexpression increased cell proliferation and expression of VEGF, VE-cadherin, and MMP2, as well as the total tube length (Fig. 4b–g). However, these effects were offset by the miR-140-3p mimic (Fig. 4b–g). MiR-140-3p mimic transfection also significantly decreased cell proliferation and expression of VEGF, VE-cadherin, and MMP2, as well as the total tube length in HUVECs (Fig. 4b–g). The results from wound-healing assay were consistent with tube formation assay (Fig. 4h, i).
SNHG1-modulated HUVEC proliferation and function by negatively regulating miR-140-3p. a RT-PCR analysis of miR-140-3p expression in HUVECs that were transfected with miR-140-3p mimic, anti-miR-140-3p, or negative controls (miR-140-3p mimic-nc or anti-nc). *P < 0.05, compared with control group. b MTT assay of cell proliferation of HUVECs that were transfected with SNHG1 and the miR-140-3p mimic of the corresponding controls (blank or miR-23a mimic-nc). The mRNA and protein levels of vascular endothelial growth factor, VE-cadherin, and MMP2 were determined by RT-PCR (c) and western blot (d), respectively. e The quantitative data of western blot. f The representative charts of tube formation using a tube formation assay. g The quantitative data of tube formation. The data were presented as mean ± SD. *P < 0.05, compared with H/R group. #P < 0.05, compared with the SNHG1 group
SNHG1 facilitated HIF-1α expression by acting as a ceRNA for miR-140-3p
The searching result of TargetScan (http://www.targetscan.org) predicted a potential binding site in the 3′-UTR of HIF-1α for miR-140-3p (Fig. 5a). We then examined whether miR-140-3p regulated HIF-1α expression in HUVECs. We found that HIF-1α expression was reduced by miR-140-3p mimic transfection and was promoted by miR-140-3p inhibitor (anti-miR-140-3p) as shown in Fig. 5b. Additionally, the result of luciferase reporter assay validated the interaction between miR-140-3p and the 3′ UTR of HIF-1α (Fig. 5c).
SNHG1 facilitated HIF-1α expression by acting as a ceRNA for miR-140-3p. a The putative miR-140-3p binding 3′ untranslated region of HIF-1α (HIF-1α-WT) and HIF-1α mutation sequence (HIF-1α-MUT). b Western blot assay and the quantitative data of the protein levels of HIF-1α in HUVECs that were transfected with miR-140-3p mimic, anti-miR-140-3p, or negative controls (miR-140-3p mimic-nc or anti-nc). *P < 0.05, compared with H/R group. c Luciferase activity assay in HUVECs. *P < 0.05, compared with mimic-nc group. d–f RNA immunoprecipitation assay was performed using anti-Ago2 in HUVECs transfected with miR-140-3p mimics or mimic-nc. SNHG1, miR-140-3p, and 3′ UTR of HIF-1α mRNA levels in the anti-Ago2-immunoprecipitated products were measured by RT-PCR. The data were presented as mean ± SD. *P < 0.05, compared with the mimic-nc group
Because miR-140-3p was predicted to bind with the same sequence shared by SNHG1 and HIF-1α, SNHG1 may act as a ceRNA for miR-140-3p to mediate HIF-1α expression in hypoxia-induced injury to vascular endothelial cells. Ago2 protein is a core component of RNA-induced silencing complex (RISC) and functions in miRNA pathway. Therefore, we carried out RIP analysis to investigate whether SNHG1, miR-140-3p, and HIF-1α are in an RISC. As shown in Fig. 5d–f, levels of SNHG1, miR-140-3p, and 3′UTR of HIF-1α mRNA in the Ago2-RIP products of miR-140-3p mimic transfected HUVECs were all significantly increased compared to the mimic-nc group (P < 0.05).
SNHG1 promoted proliferation and function of HUVECs by activating the HIF-1α/VEGF signaling pathway
Activation of HIF-1α/VEGF signaling could promote angiogenesis in ischemic tissues and exert protective function [15, 16]. VEGF expression in HUVECs significantly increased under H/R, whereas silencing HIF-1α expression with HIF-1α siRNA reduced VEGF expression under H/R (Fig. 6a, b).
SNHG1 promoted HUVECs proliferation and cell function by activating the HIF-1α and VEGF signaling pathway. a HUVECs were transfected with si-HIF-1α or corresponding controls (si-nc) under H/R. The protein expression of HIF-1α and VEGF was detected by western blot. b The quantitative data of the protein levels. *P < 0.05, compared with the control group. #P < 0.05, compared with the H/R group. c The protein levels of VEGF, VE-cadherin, and MMP2 were determined by western blot. d The quantitative data of the protein levels. e Cell proliferation was determined by MTT assay. f Tube formation was measured using a tube formation assay. g The quantitative data of tube formation. The data were presented as mean ± SD. *P < 0.05, compared with the H/R group. #P < 0.05, compared with the SNHG1 group
CBO-P11, a VEGF inhibitor, was used to determine whether SNHG1 influenced HUVEC function through VEGF signaling. After treating HUVECs with CBO-P11 (1 nM) for 48 h, the results showed that CBO-P11 suppressed the effects of SNHG1 upregulation on VEGF, VE-cadherin, and MMP2 expression (Fig. 6c, d). Also, CBO-P11 greatly decreased HUVEC proliferation, as well as tube formation (Fig. 6e–f). Collectively, these data show that SNHG1 regulated HUVEC function by activating the HIF-1α/VEGF signaling pathway.
Discussion
Multiple genes are aberrantly expressed in infarcted hearts. These genes are responsible for cardiac remodeling and cardiac injury after I/R. Formation of new blood vessels by angiogenesis and arteriogenesis is critical for restoring blood flow into ischemic heart. LncRNAs, which play vital roles in the pathogenesis of many diseases, further complicate the genetic network of angiogenesis. In this study, we found that SNHG1 expression was increased and then decreased in the cardiac microvasculature in a mouse model with cardiac I/R injury. Angiogenic activation of HUVECs is associated with increased cell proliferation and migration. Upregulation of MMP2 in HUVECs contributed markedly to cell migration. Meanwhile, various growth factors such as VEGF are involved in regulating angiogenesis. Adhesion molecule VE-cadherin is required for endothelial cell elongation during sprout outgrowth [17]. In this study, SNHG1 overexpression partly offset the effects of H/R on cell proliferation. SNHG1 upregulation also increased expression of VEGF, VE-cadherin, and MMP2. Inducing apoptosis in tube-forming endothelial cells can suppress angiogenesis [18]. We found a notable increase in capillary tubes after SNHG1 overexpression, whereas SNHG1 knockdown promoted the changes induced by H/R.
LncRNA could regulate chromatin structure and act as sponges to regulate gene expression at transcriptional and posttranscriptional level. LncRNA may function as a decoy for miRNAs, which further regulate mRNA expression though direct binding [19]. SNHG1 reportedly acts as a ceRNA to modulate derepression of miRNA targets in cell proliferation and angiogenesis. In the current study, the bioinformatics analysis revealed that miR-140-3p potentially targeted SNHG1. Previous studies have shown that miR-140 aggravates hypoxia-induced H9c2 cell injury [20], and elevates cerebral protection of dexmedetomidine against hypoxic–ischemic brain damage [21]. In this study, miR-140-3p was downregulated and then upregulated in the cardiac microvasculature of I/R injured mice. MiR-140-3p mimic transfection significantly offset the effects of SNHG1 overexpression on cell proliferation and expression of VEGF, VE-cadherin, and MMP2, as well as tube formation and wound closure in HUVECs. MiR-140-3p was confirmed to be a target of SNHG1 for the first time in the present study.
Most miRNAs conduct their biological functions by binding to target molecules [22]. In the current study, the result of luciferase reporter assay confirmed the inhibition role of miR-140-3p in regulating the transcriptional activity of HIF-1α. The result revealed that miR-140-3p may repressed HUVECs proliferation through modulating HIF-1α. HIF-1α pathway has been reported to interact with Notch signaling and engage in neuron cell death in response to ischemic stroke [23]. In the heart, HIF-1α stabilization protected cardiac cells by promoting aerobic glycolysis and reserving mitochondria function during acute ischemia–reperfusion injury [24]. The protective effect of HIF-1α on ischemic hearts is partly attributed to the induction of VEGF [25, 26]. Meanwhile, the HIF-1α/VEGF signaling pathways have been reported to be involved in an anti-ischemic stroke response by improving the microenvironment of the neurovascular unit [27]. In this study, our data showed that SNHG1 could relieve the suppression of HIF-1α expression and facilitate HIF-1α/VEGF signaling pathway transduction in promoting HUVECs survival. This process could be hindered by miR-140-3p mimic or VEGF inhibitor. It indicated that SNHG1 alleviates hypoxia-reoxygenation-induced vascular endothelial cell injury as a competing endogenous RNA through the HIF-1α/VEGF signal pathway.
Collectively, the present study demonstrated that SNHG1 protects against hypoxia-reoxygenation-induced vascular endothelial cell injury via acting as a ceRNA for miR-140-3p and activating HIF-1α/VEGF signaling pathway. It furthers understanding of cardiac I/R injury. Although LncRNA functions through multiple targets, this study sheds light into the potential of SNHG1 as a therapeutic target in cardiac I/R injury. To foster clinical translation, an in vivo study using SNHG1 transgenic mice would be desirable.
References
Moens AL, Claeys MJ, Timmermans JP, Vrints CJ (2005) Myocardial ischemia/reperfusion-injury, a clinical view on a complex pathophysiological process. Int J Cardiol 100:179–190. https://doi.org/10.1016/j.ijcard.2004.04.013
Leung A, Trac C, Jin W, Lanting L, Akbany A, Saetrom P, Schones DE, Natarajan R (2013) Novel long noncoding RNAs are regulated by angiotensin II in vascular smooth muscle cells. Circ Res 113:266–278. https://doi.org/10.1161/circresaha.112.300849
Wu P, Zuo X, Deng H, Liu X, Liu L, Ji A (2013) Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases. Brain Res Bull 97:69–80. https://doi.org/10.1016/j.brainresbull.2013.06.001
Zhang J, Yuan L, Zhang X, Hamblin MH, Zhu T, Meng F, Li Y, Chen YE, Yin KJ (2016) Altered long non-coding RNA transcriptomic profiles in brain microvascular endothelium after cerebral ischemia. Exp Neurol 277:162–170. https://doi.org/10.1016/j.expneurol.2015.12.014
Hofmann P, Sommer J, Theodorou K, Kirchhof L, Fischer A, Li Y, Perisic L, Hedin U, Maegdefessel L, Dimmeler S, Boon RA (2018) Long non-coding RNA H19 regulates endothelial cell aging via inhibition of Stat3 signaling. Cardiovasc Res. https://doi.org/10.1093/cvr/cvy206
Zhang X, Tang X, Hamblin MH, Yin KJ (2018) Long non-coding RNA Malat1 regulates angiogenesis in hindlimb ischemia. Int J Mol Sci. https://doi.org/10.3390/ijms19061723
Zhao W, Fu H, Zhang S, Sun S, Liu Y (2018) LncRNA SNHG16 drives proliferation, migration, and invasion of hemangioma endothelial cell through modulation of miR-520d-3p/STAT3 axis. Cancer Med. https://doi.org/10.1002/cam4.1562
Sun Y, Wei G, Luo H, Wu W, Skogerbo G, Luo J, Chen R (2017) The long noncoding RNA SNHG1 promotes tumor growth through regulating transcription of both local and distal genes. Oncogene 36:6774–6783. https://doi.org/10.1038/onc.2017.286
Zhang M, Wang W, Li T, Yu X, Zhu Y, Ding F, Li D, Yang T (2016) Long noncoding RNA SNHG1 predicts a poor prognosis and promotes hepatocellular carcinoma tumorigenesis. Biomed Pharmacother 80:73–79. https://doi.org/10.1016/j.biopha.2016.02.036
Zhang N, Meng X, Mei L, Hu J, Zhao C, Chen W (2018) The long non-coding RNA SNHG1 attenuates cell apoptosis by regulating miR-195 and BCL2-like protein 2 in human cardiomyocytes. Cell Physiol Biochem 50:1029–1040. https://doi.org/10.1159/000494514
Hollander MR, de Waard GA, Konijnenberg LS, Meijer-van Putten RM, van den Brom CE, Paauw N, de Vries HE, van de Ven PM, Aman J, Van Nieuw-Amerongen GP, Hordijk PL, Niessen HW, Horrevoets AJ, Van Royen N (2016) Dissecting the effects of ischemia and reperfusion on the coronary microcirculation in a rat model of acute myocardial infarction. PLoS ONE 11:e0157233. https://doi.org/10.1371/journal.pone.0157233
Ziegler T, Horstkotte M, Lange P, Ng J, Bongiovanni D, Hinkel R, Laugwitz KL, Sperandio M, Horstkotte J, Kupatt C (2016) Endothelial RAGE exacerbates acute postischaemic cardiac inflammation. Thromb Haemost 116:300–308. https://doi.org/10.1160/TH15-11-0898
Yang Q, He GW, Underwood MJ, Yu CM (2016) Cellular and molecular mechanisms of endothelial ischemia/reperfusion injury: perspectives and implications for postischemic myocardial protection. Am J Transl Res 8:765–777
Xie M, Kong Y, Tan W, May H, Battiprolu PK, Pedrozo Z, Wang ZV, Morales C, Luo X, Cho G, Jiang N, Jessen ME, Warner JJ, Lavandero S, Gillette TG, Turer AT, Hill JA (2014) Histone deacetylase inhibition blunts ischemia/reperfusion injury by inducing cardiomyocyte autophagy. Circulation 129:1139–1151. https://doi.org/10.1161/CIRCULATIONAHA.113.002416
Han MK, Kim M, Bae SY, Kang L, Han SY, Lee YS, Rha JH, Kim SU, Roh JK (2004) VEGF protects human cerebral hybrid neurons from in vitro ischemia. NeuroReport 15:847–850
Jin KL, Mao XO, Greenberg DA (2000) Vascular endothelial growth factor: direct neuroprotective effect in in vitro ischemia. Proc Natl Acad Sci USA 97:10242–10247
Betz C, Lenard A, Belting HG, Affolter M (2016) Cell behaviors and dynamics during angiogenesis. Development 143:2249–2260. https://doi.org/10.1242/dev.135616
Kunimasa K, Ahn M-R, Kobayashi T, Eguchi R, Kumazawa S, Fujimori Y, Nakano T, Nakayama T, Kaji K, Ohta T (2010) Brazilian propolis suppresses angiogenesis by inducing apoptosis in tube-forming endothelial cells through inactivation of survival signal ERK1/2. Evid Based Complement Alternat Med. https://doi.org/10.1093/ecam/nep024
Akhade VS, Pal D, Kanduri C (2017) Long noncoding rna: genome organization and mechanism of action. Adv Exp Med Biol 1008:47–74. https://doi.org/10.1007/978-981-10-5203-3_2
Xing B, Li QJ, Li H, Chen SS, Cui ZY, Ma J, Zhang ZW (2018) miR-140-5p aggravates hypoxia-induced cell injury via regulating MLK3 in H9c2 cells. Biomed Pharmacother 103:1652–1657. https://doi.org/10.1016/j.biopha.2018.04.062
Han XR, Wen X, Wang YJ, Wang S, Shen M, Zhang ZF, Fan SH, Shan Q, Wang L, Li MQ, Hu B, Sun CH, Wu DM, Lu J, Zheng YL (2018) MicroRNA-140-5p elevates cerebral protection of dexmedetomidine against hypoxic-ischaemic brain damage via the Wnt/beta-catenin signalling pathway. J Cell Mol Med 22:3167–3182. https://doi.org/10.1111/jcmm.13597
Ebert MS, Sharp PA (2012) Roles for microRNAs in conferring robustness to biological processes. Cell 149:515–524
Cheng YL, Park JS, Manzanero S, Choi Y, Baik SH, Okun E, Gelderblom M, Fann DY, Magnus T, Launikonis BS, Mattson MP, Sobey CG, Jo DG, Arumugam TV (2014) Evidence that collaboration between HIF-1alpha and Notch-1 promotes neuronal cell death in ischemic stroke. Neurobiol Dis 62:286–295. https://doi.org/10.1016/j.nbd.2013.10.009
Ong SG, Lee WH, Theodorou L, Kodo K, Lim SY, Shukla DH, Briston T, Kiriakidis S, Ashcroft M, Davidson SM, Maxwell PH, Yellon DM, Hausenloy DJ (2014) HIF-1 reduces ischaemia-reperfusion injury in the heart by targeting the mitochondrial permeability transition pore. Cardiovasc Res 104:24–36. https://doi.org/10.1093/cvr/cvu172
Du Y, Ge Y, Xu Z, Aa N, Gu X, Meng H, Lin Z, Zhu D, Shi J, Zhuang R, Wu X, Wang X, Yang Z (2018) Hypoxia-inducible factor 1 alpha (HIF-1alpha)/vascular endothelial growth factor (VEGF) pathway participates in angiogenesis of myocardial infarction in muscone-treated mice: preliminary study. Med Sci Monit 24:8870–8877. https://doi.org/10.12659/MSM.912051
He Y, Yu S, Hu J, Cui Y, Liu P (2016) Changes in the anatomic and microscopic structure and the expression of HIF-1alpha and VEGF of the yak heart with aging and hypoxia. PLoS ONE 11:e0149947. https://doi.org/10.1371/journal.pone.0149947
Wu C, Chen J, Chen C, Wang W, Wen L, Gao K, Chen X, Xiong S, Zhao H, Li S (2015) Wnt/beta-catenin coupled with HIF-1alpha/VEGF signaling pathways involved in galangin neurovascular unit protection from focal cerebral ischemia. Sci Rep 5:16151. https://doi.org/10.1038/srep16151
Acknowledgements
None.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared that no competing interests exist.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liang, S., Ren, K., Li, B. et al. LncRNA SNHG1 alleviates hypoxia-reoxygenation-induced vascular endothelial cell injury as a competing endogenous RNA through the HIF-1α/VEGF signal pathway. Mol Cell Biochem 465, 1–11 (2020). https://doi.org/10.1007/s11010-019-03662-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-019-03662-0