Abstract
Ovarian cancer (OC) is the most lethal gynecologic malignancy and long non-coding RNAs (lncRNAs) have been acknowledged as important regulators in human OC. This study aimed to investigate the function and underlying mechanisms of LINC00504 in OC. The expression levels of LINC00504 in human OC tissues and cell lines were investigated by qRT-PCR analysis. The OC cell proliferation, and apoptosis were evaluated by MTT assay, colony-formation assay, Caspase-3 activity assay, and nucleosome ELISA assay, respectively. The metabolic shift in OC cells was examined by aerobic glycolysis analysis. Dual-luciferase activity reporter assay and mRNA–miRNA pull-down assay were conducted to validate the interaction between LINC00504 and miR-1244. LINC00504 was upregulated in OC cell lines and specimens. Knockdown of LINC00504 inhibited cell proliferation, enhanced apoptosis, decreased glycolysis-related gene (PKM2, HK2, and PDK1) expression, and altered aerobic glycolysis in OC cells and vice versa. LINC00504 downregulated miR-1244 expression levels by acting as an endogenous sponge of miR-1244. Inhibition of miR-1244 diminished the effects of LINC00504 on OC cells. Our study shows that LINC00504 promotes OC cell progression and stimulates aerobic glycolysis by interacting with miR-1244, which indicates that LINC00504 might act as a promising therapeutic target for OC treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Ovarian cancer (OC) is the second most common and the most lethal gynecologic malignancy in the western world [1]. OC ranks the fifth in the cause of cancer-related death with an estimated 240,000 new cases and 160,000 deaths worldwide annually [2,3,4]. Due to the absence of early warning symptoms and deficiency in screening programs, around 70% of OC cases are diagnosed at the advanced stage and metastasizes to other organs, resulting in bad prognosis and high mortality rates [5,6,7]. Despite the development of advanced diagnosis and surgical technologies, as well as many new anti-cancer drugs, the mortality rate of OC has been only modestly changed over the last decade [8,9,10]. Thus, a better understanding of the molecular mechanisms underlying the development and progression of OC is urgently needed.
In the recent years, a growing body of researches illustrated an intriguing correlation between long non-coding RNAs (lncRNAs) and human cancers. LncRNAs are non-protein-coding RNAs with a length of more than 200 nucleotides [11,12,13]. For decades, these lncRNAs were recognized at transcriptional “noise”. With the development of transcriptomic sequencing technologies and high-resolution microarray, the importance of lncRNAs are gradually gain attention [14, 15]. LncRNAs are reported to participate in many fundamental functions of cells, including histone modification, modification of alternative splicing genes, chromatin structure modification, and miRNAs expression regulation. Thus, lncRNAs play crucial roles in genomic imprinting, cell differentiation, organogenesis, and tumorigenesis [16,17,18,19,20]. Currently, only a small proportion of lncRNAs have been functionally characterized and the majority of lncRNA function, especially in OC, remains elusive.
Our pilot results clearly demonstrated that the expression levels of LINC00504 were dramatically upregulated in human OC specimens and cell lines. In this study, we aimed to investigate the biological function roles of LINC00504 in OC. We intended to address several important questions: (1) What is the effect of LINC00504 on OC cell proliferation, colony formation, and apoptosis. (2) What is the effect of LINC00504 on the metabolic changes and the metabolic genes alternation in OC cells. (3) What is the potential direct target of LINC00504 that may be associated with its function. The answer to these questions would provide new insights into the functional roles of LINC00504 in OC development as well as may beneficial for the development of new therapeutic approaches for the treatment of OC patients.
Materials and methods
Patients’ specimens
OC tissues and matched adjacent normal tissues were obtained from 45 patients, including 23 stage 1–2 patients and 22 stage 3–4 patients. All the histological diagnoses for OC and normal tissues were reviewed and recognized by two pathologists independently. All patients’ stages were based on the endometrial and OC-staging criteria set by the International Federation of Gynecology and Obstetrics (FIGO). Written informed consent was obtained from each participant prior to sample collection. The study protocol was approved by the Ethics Committee of Heze Municipal Hospital. The human specimens were immediately frozen in liquid nitrogen and stored at − 80 °C before use.
Cell culture
HOSEpiC (the normal ovarian surface epithelial cell line) and five human OC cell lines A2780, CAOV-3, HO-8910, OVCAR-3 and SKOV-3 were obtained from the American Type Culture Collection (ATCC, Manassas, USA) and cultured according to the provider’s instructions.
Quantitative real-time PCR (qRT-PCR)
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, USA). First-strand cDNA was synthesized using a First-Strand cDNA synthesis kit (OriGene Technologies, Rockville, USA). Amplifications were performed with Brilliant SYBR® II QPCR Master kit (Agilent Technologies, Santa Clara, USA) on a ViiA7 real-time PCR system (Applied Biosystems, Foster City, USA). Following the manufacturer’s instructions, reactions were carried out in a final volume of 25 μl with 1.5 μl of cDNA and 1 μl of each primer at the optimized concentration. The relative LINC00504 expression was normalized to β-actin, which was calculated using the \(2^{{ - \Delta \Delta C_{\text{t}} }}\) method. The primers used in this study are as follows: LINC00504, the forward primer (5′-ATACAGTCAAAGAGCGGATGGCC-3′) and the reverse primer (5′-GGTACTTTCACACTGGACTG-3′); Pyruvate kinase isozymes M2 (PKM2), the forward primer (5′-CCACTTGCAATTATTTGAGGAA-3′) and the reverse primer (5′-GTGAGCAGACCTGCCAGACT-3′); Hexokinase-2 (HK2), the forward primer (5′-TGTGCGTAATGGCAAGCGGAGG-3′) and the reverse primer (5′-CCACGGCAACCACATCCAGGTC-3′); Pyruvate Dehydrogenase Kinase 1 (PDK1), the forward primer (5′-GTGTAGATTAGAGGGATG-3′) and the reverse primer (5′-AAGGAATAGTGGGTTAGG-3′); β-actin, the forward primer (5′-GCACCGTCAAGGCTGAGAAT-3′) and the reverse primer (5′-GGTGAAGACGCCAGTGGA-3′); miR-1244, reverse transcription primer:5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACAACCAT-’3. The forward primer (5′-TTACAAAGTAGTTGGTTTGTATGAG-3′) and the reverse primer (5′-TGGTGTCGTGGAGTCG-3′); U6, reverse transcription primer 5′-AACGCTTCACGAATTTGCGT-3′, the forward primers (5′-CTCGCTTCGGCAGCACA-3′) and the reverse primer (5′-TGGTGTCGTGGAGTCG-3′).
Silencing RNAs (siRNAs) or miRNAs
OVCAR-3 and SKOV-3 human OC cells were seeded in six-well plates and grown to 50% confluence. After 24 h, the cells were transfected with siRNA using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer’s instructions. The two LINC00504 mRNA target sequences were si-LINC00504#1: 5′-GAGAGAAGAATCAAATAGACT-3′, si-LINC00504#2: 5′-GAGAAGAATCAAATAGACTCA-3′. miR-NC and miR-1244 precursor were purchased from ThomasFisher Scientific (Walthma, USA).
MTT assay
OVCAR-3 and SKOV-3 cells were seeded into 96-well plates (2 × 103 cells/well) and incubated for 0, 24, 48, 72, 96, and 120 h. Then 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) (5 mg/ml) was added to each well. After 4 h, the formed formazan crystals were dissolved in DMSO (150 μl) and the absorbance was measured at 490 nm using a microplate spectrophotometer (Thermo Spectronic, Madison, USA).
Colony-formation assay
OVCAR-3 and SKOV-3 cells were seeded into in 6-well tissue culture plates (200 cells/well). On the second day, cells were transfected with the LINC00504 siRNAs or negative control siRNAs. 10 days later the colonies were fixed in ethanol and stained with 0.1% crystal violet. Colonies (> 50 cells or 1 mm in diameter) were counted by two individuals.
Caspase-3 activity assay
Caspase-3 activity was measured using Caspase-Glo 3/7 Assay (Promega, Madison, USA) following the manufacturer’s instructions. OVCAR-3 and SKOV-3 Cells were grown in 96-well plates, 100 µl Caspase-Glo reagent was added to each well and samples were incubated for 30 min at room temperature with constant shaking. Enzyme activity was measured using a GENiosPro plate reader.
Nucleosome ELISA assay
Nucleosome ELISA was done using a Nucleosome ELISA kit from Calbiochem (San Diego, USA) according to the manufacturer’s protocol. Briefly, OVCAR-3 and SKOV-3 cells were seeded in 100 mm culture dishes. After transfection with the LINC00504 siRNAs or negative control siRNAs for 48 h, cells were harvested, and nuclear lysates were prepared. A 1:5 dilution of lysate was added into wells of a pretreated ELISA plate. The plate was then incubated with detector antibody followed by streptavidin conjugate. After washing, substrate solution was added, and plates were incubated in the dark for 30 min. The reaction was stopped with stop solution, and absorbance was measured on an ELISA reader at a test wavelength of 450 nm.
Analysis of lactate production, glucose uptake, ATP level and oxygen consumption
OVCAR-3 and SKOV-3 cells were transfected with the LINC00504 siRNAs or negative control siRNAs. Lactate production was measured by the lactate assay kit (BioVision, Mountain View, USA) following the manufacturer’s protocol. Glucose uptake was measured using glucose uptake cell-based assay kit (Cayman Chemicals, Michigan, USA) according to manufacturer’s instruction. ATP levels were tested by a CellTiter-Glo® luminescent cell viability assay kit (Promega, Madison, USA) following the manufacturer’s protocol. Oxygen consumption was performed using the SeaHorse XF24 extracellular analyzer (SeaHorse Bioscience, Billerica, USA) by 8-min cyclic protocol commands (mix for 3 min, let stand 2 min and measure for 3 min).
Western blot
Cells were harvested and lysed with RIPA buffer containing the protease inhibitors. The total protein concentration was determined by the bicinchoninic acid (BCA) assay. Equal amounts of protein were loaded to 10% SDS–polyacrylamide gel electrophoresis and separated by electrophoresis. Then, the protein bands were transferred onto polyvinylidene difluoride (PVDF, Bio-Rad, Hercules, USA) membranes which were blocked with 5% milk for 1 h at room temperature, subsequently followed by incubating with primary antibodies against PKM2, HK2 and PDK1 (Cell Signaling Technology, Danvers, USA) overnight at 4 °C. Next, the membranes were incubated with HRP-conjugated secondary antibodies for 1 h at room temperature. Signals were visualized using ECL (Millipore, Burlington, USA) according to the manufacturer’s protocol.
Luciferase reporter assay
OVCAR-3 and SKOV-3 cells were seeded 24-well plates. On the following day, cells were co-transfected with miR-1244 or miR-NC mimics and luciferase reporters containing WT-LINC00504 or Mut-LINC00504. The pRT-TK Renilla plasmid was also co-transfected. Luciferase activity was measured using the Dual-Luciferase reporter assay system (Promega, WI, USA) and the data were detected using a GloMax 20/20 luminometer (Promega) according to manufacturer’s instruction. Mutations in the putative binding sites of LINC00504 were made using Quikchange site-directed mutagenesis kit (Agilent Technologies, Santa Clara, USA). The primers were as following: WT-LINC00504, Forward primer: 5′- GTGACTCGAGCTTGCCTCTGCCATGT-3′, Reverse primer: 5′-GTGAGCGGCCGC TTTCAGAGTGAAACAATACTT-3′. Mut-LINC00504, Forward primer: 5′-TCTTCAAGGAGAACTCCCACCAAGTCCA-3′, Reverse primer: 5′-TGGACTTGGTGGGAGTTCTCCTTGAAGA-3′.
Biotinylated miRNA pull-down assay
Lysates from OVCAR-3 and SKOV-3 cells were incubated with in vitro-synthesized biotin-labeled sense or antisense DNA probes against LINC00504 for 1 h. Dynabeads M-280 Streptavidin beads (Life Technologies, Carlsbad, USA) were blocked for 2 h and added to the cell lysis to pull down the biotin-coupled RNA–RNA complex. LINC00504 and miR-1244 levels were analyzed by qRT-PCR. DNA probes are as follows: LINC00504-DNA-1-sense: biotin-GCAACACATCAAAAAGCTTATCCACCATGA, LINC00504-DNA-1-antisense: biotin-TCATGGTGGATAAGCTTTTTGATGTGTTGC; LINC00504-DNA-2-sense: biotin-GTTTGCAGATGACATGATTGTATATCTAGA, LINC00504-DNA-2-antisense: biotin-TCTAGATATACAATCATGTCATCTGCAAAC; LINC00504-DNA-3-sense: biotin-GGAACATCACACTCTGGGGACTGTTGTGGG, LINC00504-DNA-3-antisense: biotin-CCCACAACAGTCCCCAGAGTGTGATGTTCC.
Statistical analysis
Results are expressed as the mean ± SD of three independent experiments. For two-group comparison analysis, the statistical significance of differences between means was analyzed by unpaired Student’s t test. For multiple group comparison analysis, one- or two-way ANOVA with Bonferroni post hoc test was employed. P < 0.05 were considered statistically significant. The survival rate was estimated by the Kaplan–Meier method, with statistical significance set at 0.05 and determined using the log-rank test.
Results
LINC00504 are upregulated in OC tumor tissues and cell lines
The expression levels of LINC00504 in 45 paired OC tissues and matched adjacent noncancerous normal tissues were determined by qRT-PCR. We found that 33 out of 45 samples showed significant upregulation of the levels of LINC00504 in OC tissues compared with that in normal tissues (Fig. 1a). Moreover, the levels of LINC00504 were higher in the late stage (3–4) than that in the early stage (1–2) of OC tissues (Fig. 1b). More importantly, the OC patients with LINC00504-high exhibited a worse overall survival rate compared with that in OC patients with LINC00504-low (Fig. 1c). We further analyzed the expression profile of LINC00504 in five human ovarian cancer cell lines (A2780, CAOV-3, HO-8910, OVCAR-3, and SKOV-3) and one normal ovarian surface epithelial cell line (HOSEpiC). Consistent with the results from human OC and normal tissues, the expression levels of LINC00504 were also markedly upregulated in all five OC cell lines compared to HOSEpiC cells (Fig. 1d). These results suggested that LINC00504 was significantly upregulated in both OC tissues and cell lines, indicating that LINC00504 may play an essential role in the development of OC.
LINC00504 was up-regulated in ovarian cancer and predicted poor prognosis. a The RNA levels of LINC00504 in ovarian cancer tissues and pair-matched adjacent noncancerous normal tissues were analyzed by qRT-PCR (shown as \(2^{{ - \Delta \Delta C_{\text{t}} }}\)). b qRT-PCR showed higher LINC00504 levels were associated with advanced TNM stages. c Overall survival curve was plotted by the Kaplan–Meier method, according to LINC00504 expression levels. d LINC00504 expression was measured in five human ovarian cancer cell lines (A2780, CAOV-3, HO-8910, OVCAR-3 and SKOV-3) and the normal ovarian surface epithelial cell line (HOSEpiC) by qRT-PCR. *P < 0.05, **P < 0.01
LINC00504 plays an oncogenic role in OC cells
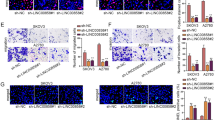
To investigate the biological function of LINC00504, two OC cell lines (OVCAR-3 and SKOV-3) were selected for LINC00504 knockdown experiments due to their high endogenous levels of LINC00504. OVCAR-3 and SKOV-3 were transfected with two si-LINC00504 specific designed to target LINC00504. The qRT-PCR results showed that transfection of either si-LINC00504-1 or si-LINC00504-2 resulted in more than 70–80% of reduction of LINC00504 levels in both OVCAR-3 and SKOV-3 cells (Fig. 2a). We found that knockdown of LINC00504 by either si-LINC00504-1 or si-LINC00504-2 significantly decreased cell proliferation of OC as demonstrated by MTT assay and colony-formation assay (Fig. 2b–d). Furthermore, we observed that the caspase-3 activity and nucleosome content were dramatically upregulated in LINC00504 knockdown OC cells compared to nonsense-control (NC)-transfected cells (Fig. 2e, f). Collectively, these results suggested that LINC00504 knockdown inhibited cell proliferation and promoted cell apoptosis of OC cells, indicating that LINC00504 may play an oncogenic role in OC cells.
Knockdown of LINC00504 inhibited proliferation and promoted apoptosis of ovarian cancer cells. a qRT-PCR analysis of LINC00504 levels in OVCAR-3 and SKOV-3 cells transfected with LINC00504 siRNAs (si-LINC00504#1, si-LINC00504#2) or negative control (si-NC). b, c The proliferation of OVCAR-3 (b) and SKOV-3 (c) cells transfected with LINC00504 siRNAs (si-LINC00504#1, si-LINC00504#2) or negative control (si-NC) was determined by MTT assay. d Colony formation ability was evaluated by colony formation assay in OVCAR-3 and SKOV-3 cells transfected with LINC00504 siRNAs (si-LINC00504#1, si-LINC00504#2) or negative control (si-NC). e, f Apoptotic rate of OVCAR-3 and SKOV-3 cells transfected with LINC00504 siRNAs (si-LINC00504#1, si-LINC00504#2) or negative control (si-NC) was measured by caspase 3 activity assay (e) and nucleosome ELISA assay (f). The data represent the mean ± SD from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001
Knockdown of LINC00504 suppresses aerobic glycolysis in OC cells
To investigate the effect of LINC00504 on the metabolic phenotype of OC cell lines, OVCAR-3 and SKOV-3 were transfected with two si-LINC00504 − 1 or − 2 to knockdown of LINC00504. We performed aerobic glycolysis analysis using conditioned media collected from these OC cell lines for glucose consumption, lactate, and ATP production analysis. We found that the glucose consumption, lactate, and ATP production levels were significantly decreased in OC cells with LINC00504-knockdown compared with those in the control cells with NC transfection (Fig. 3a–c). The suppression of the aerobic glycolytic phenotype was accompanied by increasing O2 consumption in OC cells with LINC00504-knockdown, suggesting LINC00504 depletion promote a metabolic shift towards mitochondrial respiration (Fig. 3d). These metabolic genes including PKM2, HK2, and PDK1 are known to play crucial roles in the promotion of aerobic glycolysis in cancer cells. Knockdown of LINC00504 resulted in reduction of both mRNA and protein levels of PKM2, HK2, and PDK1 in OVCAR-3 and SKOV-3 cell lines (Fig. 3e–g).
LINC00504 regulated the metabolic phenotype of ovarian cancer cells. OVCAR-3 and SKOV-3 cells transfected with LINC00504 siRNAs (si-LINC00504#1, si-LINC00504#2) or negative control (si-NC). a The lactate production was measured by the lactate assay kit. b The glucose uptake was measured by glucose uptake cell-based assay kit. c ATP level changes was measured by CellTiter-Glo luminescent cell viability assay kit. d Oxygen consumption was measured by SeaHorse XF24 extracellular analyzer. e, f The mRNA levels of glycolysis-related genes (PKM2, HK2 and PDK1) in OVCAR-3 and SKOV-3 were measured by qRT-PCR. g The mRNA levels of glycolysis-related genes (PKM2, HK2 and PDK1) in OVCAR-3 and SKOV-3 were measured Western blot. The data represent the mean ± SD from three independent experiments. *P < 0.05, **P < 0.01
Reciprocal relationship of LINC00504 and miR-1244
Previous studies demonstrated that lncRNAs can be regulated by miRNAs [21, 22]. To search the potential miRNA that can target LINC00504, we applied a well-developed online lncRNAs target searching algorithm-miRcode (http://www.mircode.org/mircode/) and we identified that miR-1244 is a potential target of LINC00504. Using RNAhybrid algorithm (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid), we further identified the putative binding site of LINC00504 for miR-1244 binding (Fig. 4a). To experimentally validate this finding, we constructed two luciferase reporter plasmids containing the wide-type (WT) or mutated (Mut) binding site of LINC00504. Overexpression of miR-1244 significantly decreased the wild-type LINC00504 luciferase activity, whereas the mutant LINC00504 reporter luciferase activity was not affected in OVCAR-3 and SKOV-3 cell lines (Fig. 4b, c). Furthermore, the biotin-tagged LINC00504 was used for RNA immunoprecipitation assay. The qRT-PCR results showed that LINC00504 can efficiently bind with miR-1244 and both LINC00504 and miR-1244 were enriched in RNA immunoprecipitation complex (Fig. 4d, e). Interestingly, knockdown of LINC00504 led to 5.8–7.5 folds upregulation of miR-1244 in OVCAR-3 and SKOV-3 cell lines (Fig. 4f). Finally, an inverse correlation was observed between miR-1244 and LINC00504 in 45 OC tissue samples (Fig. 4g). These results suggested that a reciprocal relationship exists between miR-1244 and LINC00504, and LINC00504 acts as a sponge for miR-1244.
LINC00504 acted as a sponge of miR-1244 in ovarian cancer cells. a The predicted wild-type (WT-LINC00504) and mutated (Mut-LINC00504) miR-1244 binding sites in LINC00504 transcript were shown. b, c The luciferase activities in OVCAR-3 and SKOV-3 cells co-transfected with miR-1244 or miR-NC mimics and luciferase reporters containing WT-LINC00504 or Mut-LINC00504 were measured using the Dual-Luciferase reporter assay system. d, e Lysates from OVCAR-3 and SKOV-3 cells were incubated with in vitro-synthesized biotin-labeled sense or antisense DNA probes against LINC00504 for biotin pull-down assay, followed by qRT-PCR analysis to examine LINC00504 and miR-1244 levels. f qRT-PCR analysis of miR-1244 levels in OVCAR-3 and SKOV-3 cells transfected with LINC00504 siRNAs (si-LINC00504#1, si-LINC00504#2) or negative control (si-NC). g There was an inverse expression correlation between LINC00504 and miR-1244 in 45 ovarian cancer tissues. *P < 0.05, **P < 0.01. ns not significant
LINC00504 regulates OC cell function through mediating miR-1244
Based on our findings, we hypnotized that the functional effect of LINC00504 on OC cell proliferation, apoptosis, and aerobic glycolysis may through the regulation of miR-1244. To verify this hypothesis, we first aimed to knockdown of miR-1244 in OC cell lines with LINC00504 silencing. Because Fig. 4f showed that knockdown of LINC00504 upregulated miR-1244. SKOV-3 cells were co-transfected with si-NC + NC-inhibitor, si-LINC00504-1 + NC-inhibitor, or si-LINC00504-1 + miR-1244-inhibitor. The qRT-PCR results showed that miR-1244-inhibitor completely abolished the effect of LINC00504-1 knockdown-induced miR-1244 upregulation (Fig. 5a). Interestingly, we observed that miR-1244-inhibitor transfection completely restored the inhibition effect of LINC00504-1 knockdown-mediated SKOV-3 cell proliferation (Fig. 5b), colony formation (Fig. 5c), and glucose consumption, lactate, and ATP production (Fig. 5f–h). Suppression of miR-1244 also rescued the expression of both mRNA and protein levels of PKM2, HK2, and PDK1 in and SKOV-3 cell lines with LINC00504-1 knockdown (Fig. 5g, h, J,k). Finally, we found that the effect of LINC00504-1 knockdown-induced cell apoptosis and O2 consumption of SKOV-3 was blocked via decreasing of miR-1244 (Fig. 5d, i).
MiR-1244 inhibition significantly rescued the decreased proliferation and aerobic glycolysis due to knockdown of LINC00504 in ovarian cancer cells. a The miR-1244 expression was measured in SKOV-3 cells co-transfected with si-NC and NC-inhibitor, si-LINC00505#1 and NC-inhibitor, or si-LINC00505#1 and miR-1244-inhibitor by qRT-PCR. b, c The proliferation of SKOV-3 cells co-transfected with si-NC and NC-inhibitor, si-LINC00505#1 and NC-inhibitor, or si-LINC00505#1 and miR-1244-inhibitor was determined by MTT assay (b) and colony formation assay (c). d, e Apoptotic rate of SKOV-3 cells co-transfected with si-NC and NC-inhibitor, si-LINC00505#1 and NC-inhibitor, or si-LINC00505#1 and miR-1244-inhibitor was measured by caspase 3 activity assay (d) and nucleosome ELISA assay (e). f–h MiR-1244 inhibition rescued the inhibition effect on lactate production, glucose uptake and ATP level due to knockdown of LINC00505 in SKOV-3 cells. i MiR-1244 inhibition impaired the promotion effect on oxygen consumption due to knockdown of LINC00505 in SKOV-3 cells. j, k MiR-1244 inhibition rescued the suppression effect on the expression of glycolysis-related genes (PKM2, HK2 and PDK1) due to knockdown of LINC00505 in SKOV-3 cells. The data represent the mean ± SD from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001
To further verify our hypothesis, SKOV-3 cells were co-transfected with Vector + miR-NC, LINC00504-(overexpression) OE + miR-NC, or LINC00504-OE + miR-1244. Forced expression of LINC00504 evidently reduced miR-1244 expression, and this inhibition effect can be rescued by overexpression of miR-1244 in SKOV-3 (Fig. 6a). We found that upregulation of LINC00504 promoted SKOV-3 cell proliferation (Fig. 6b), colony formation (Fig. 6c), potential survival ability (Fig. 6d) and glucose consumption, lactate, and ATP production (Fig. 6g–h) as well as decreased O2 consumption (Fig. 6i). LINC00504 overexpression also led to upregulation of both mRNA and protein levels of PKM2, HK2, and PDK1 in and SKOV-3 cell lines (Fig. 6j, k). Importantly, LINC00504-mediated promotion effects on SKOV-3 cell growth and metabolism can be abolished via restored expression of miR-1244.
MiR-1244 significantly attenuated the increased proliferation and aerobic glycolysis due to overexpression of LINC00504 in ovarian cancer cells. a The miR-1244 expression was measured in SKOV-3 cells co-transfected with empty vector and miR-NC, LINC00504 overexpression plasmid and miR-NC, or LINC00504 overexpression plasmid and miR-1244 by qRT-PCR. b, c The proliferation of SKOV-3 cells co-transfected with empty vector and miR-NC, LINC00504 overexpression plasmid and miR-NC, or LINC00504 overexpression plasmid and miR-1244 was determined by MTT assay (b) and colony formation assay (c). d, e Apoptotic rate of SKOV-3 cells co-transfected with empty vector and miR-NC, LINC00504 overexpression plasmid and miR-NC, or LINC00504 overexpression plasmid and miR-1244 was measured by caspase 3 activity assay (d) and nucleosome ELISA assay (e). f–h MiR-1244 attenuated the enhanced effect on lactate production, glucose uptake and ATP level due to overexpression of LINC00505 in SKOV-3 cells. i MiR-1244 inhibition rescued the inhibition effect on oxygen consumption due to overexpression of LINC00505 in SKOV-3 cells. j, k MiR-1244 impaired the promotion effect on the expression of glycolysis-related genes (PKM2, HK2 and PDK1) due to overexpression of LINC00505 in SKOV-3 cells. The data represent the mean ± SD from three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001
Collectively, these results suggested that LINC00504 regulates OC cell proliferation, apoptosis, and aerobic glycolysis through affecting miR-1244
Discussion
Non-coding RNAs (ncRNAs) are RNA molecules transcribed from the genome that do not encode proteins, but they were shown to have specific functions. ncRNAs can be divided into two main types according to their size: small non-coding RNA (sncRNAs) (≤ 200 nucleotides), including microRNA, small interfering RNAs (siRNAs), and small nucleolar RNA (snoRNA), and lncRNAs (> 200 nucleotides) [23, 24]. While the biological functions of sncRNAs have been intensely investigated for many years on numerous human diseases initiation and progression, the functional relevance of lncRNAs is relatively less known.
Accumulating evidence has suggested that lncRNAs are involved in many biological and cellular aspects, such as cell proliferation, differentiation, and apoptosis, indicating fundamental roles for lncRNAs in tumorigenesis processes [16, 18]. The expression pattern of lncRNAs in OC have been assessed by several groups. Liu et al. reported that among 4956 lncRNAs being analyzed, 583 and 578 were upregulated and downregulated in highly metastatic OC cells, compared to the parental OC cells, respectively [25]. Besides, Qiu et al. showed that 115 lncRNAs are significantly changed in estrogen-treated OC cells compared to untreated controls, and they further revealed that upregulated TC0101441 and downregulated TC0100223 and TC0101686 are correlated with the status of estrogen receptor in OC tissues [26]. In our current study, we demonstrated that the expression levels of LINC00504 were significantly upregulated in OC tissues and cells when compared with their counterparts, respectively. These results suggested that LINC00504 may play an oncogenic role in the development of OC.
The function for some of lncRNAs in the regulation of gene expression in OC has also been elaborated. For example, AB073614 has been shown to be upregulated in OC tissues, and high levels of AB073614 are correlated with the poor overall survival rate of OC patients. AB073614 exerted an oncogenic effect on OC cells through regulation of extracellular signal-regulated kinase (ERK) 1/2 and Protein Kinase B Alpha (PKB, AKT)-mediated signaling pathway [27]. Growth arrest-specific transcript 5 (GAS5) was downregulated in epithelial OC tissues compared to normal ovarian epithelial tissues. Restoration of GAS5 suppressed cell proliferation, decreased cell migration and invasion, as well as induced apoptosis of OC cells [28,29,30]. A recent publication from Fang et al. elaborated revealed that LINC00504 regulates tumor metabolism through interaction with c-Myc in colon cancer [31]. Our results showed that knockdown of LINC00504 dramatically inhibited cell proliferation, decreased aerobic glycolysis, and promoted cell apoptosis of OC cells. These results confirmed our hypothesis that LINC00504 is an oncogene in OC cells. Using the gene-deficient mice would be beneficial for characterization of the role of LINC00504, however, we would do systematic research using in vivo mouse model in the later work.
Although our data indicated that LINC00504 exerts a tumor-promotion effect on OC cells, the underlying molecular mechanism was not fully understood. LncRNAs possess secondary structures which facilitate the interactions with DNA and RNA. Thus, lncRNAs generally regulate gene expression via binding to DNA or RNA in a sequence-specific manner [32,33,34]. Several studies reported that some lncRNAs affect target proteins expression through interaction with miRNAs. For example, lncRNA HOST2 binds with let-7b, a well-known tumor suppressor, to promoter the endogenous levels of metastasis-promoting genes including High Mobility Group AT-Hook 2 (HMGA2), c-Myc, Dicer 1, and IMP U3 Small Nucleolar Ribonucleoprotein 3 (Imp3) that were targeted by let-7b [35]. MALAT1 acted as an oncogenic lncRNA via binding with miR-506 and further regulated Protein Phosphatase 1 Regulatory Subunit 13 Like (PPP1R13L) gene expression to promote OC cell proliferation [36]. In our current study, we identified that miR-1244 is a direct target of LINC00504. Knockdown of LINC00504 enhanced miR-1244 expression and vice versa, indicating a reciprocal relationship of LINC00504 and miR-1244 exists in OC cells. MiR-1244 was reported to play a tumor suppressor role in lung cancer cells. However, the role of miR-1244 in OC cells is still unknown. Our results in Figs. 5 and 6 also suggested that miR-1244 acts as a tumor suppressor in OC cells. Our further studies will focus on the investigation of the gene(s) or signaling pathway(s) that regulated by miR-1244, which also involved in LINC00504-mediated function of OC cells.
Conclusions
Our current study, for the first time, revealed the biological functional role of LINC00504 in OC, which is that LINC00504 plays an oncogenic role in the promotion of OC progression through interaction with miR-1244. Targeting of LINC00504 or miR-1244 may provide a new therapeutic treatment for OC patients.
References
Bray F et al (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68(6):394–424
Rauh-Hain JA et al (2011) Ovarian cancer screening and early detection in the general population. Rev Obstet Gynecol 4(1):15–21
Reid BM, Permuth JB, Sellers TA (2017) Epidemiology of ovarian cancer: a review. Cancer Biol Med 14(1):9–32
Desai A et al (2014) Epithelial ovarian cancer: an overview. World J Transl Med 3(1):1–8
Doubeni CA, Doubeni AR, Myers AE (2016) Diagnosis and management of ovarian cancer. Am Fam Physician 93(11):937–944
Orr B, Edwards RP (2018) Diagnosis and treatment of ovarian cancer. Hematol Oncol Clin North Am 32(6):943–964
Bhoola S, Hoskins WJ (2006) Diagnosis and management of epithelial ovarian cancer. Obstet Gynecol 107(6):1399–1410
Lee JY et al (2018) Changes in ovarian cancer survival during the 20 years before the era of targeted therapy. BMC Cancer 18(1):601
Torre LA et al (2018) Ovarian cancer statistics, 2018. CA Cancer J Clin 68(4):284–296
Razi S et al (2016) The incidence and mortality of ovarian cancer and their relationship with the Human Development Index in Asia. Ecancermedicalscience 10:628
Thin KZ et al (2018) LncRNA-DANCR: a valuable cancer related long non-coding RNA for human cancers. Pathol Res Pract 214(6):801–805
Schmitt AM, Chang HY (2016) Long noncoding RNAs in cancer pathways. Cancer Cell 29(4):452–463
Ji D et al (2018) The role of long non-coding RNA AFAP1-AS1 in human malignant tumors. Pathol Res Pract 214(10):1524–1531
Dianatpour A, Ghafouri-Fard S (2017) Long non coding RNA expression intersecting cancer and spermatogenesis: a systematic review. Asian Pac J Cancer Prev 18(10):2601–2610
Zampetaki A, Albrecht A, Steinhofel K (2018) Long non-coding RNA structure and function: is there a link? Front Physiol 9:1201
Fang Y, Fullwood MJ (2016) Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinform 14(1):42–54
Wang KC, Chang HY (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell 43(6):904–914
Cao J (2014) The functional role of long non-coding RNAs and epigenetics. Biol Proced Online 16:11
Mattick JS, Makunin IV (2006) Non-coding RNA. Hum Mol Genet 15(1):R17–R29
Yang JX, Rastetter RH, Wilhelm D (2016) Non-coding RNAs: an introduction. Adv Exp Med Biol 886:13–32
Paraskevopoulou MD, Hatzigeorgiou AG (2016) Analyzing miRNA–LncRNA interactions. Methods Mol Biol 1402:271–286
Zhang Y et al (2017) Decoding noncoding RNAs: role of microRNAs and long noncoding RNAs in ocular neovascularization. Theranostics 7(12):3155–3167
Hube F, Francastel C (2018) Coding and non-coding RNAs, the frontier has never been so blurred. Front Genet 9:140
Huang B, Zhang R (2014) Regulatory non-coding RNAs: revolutionizing the RNA world. Mol Biol Rep 41(6):3915–3923
Liu SP et al (2013) Identification of differentially expressed long non-coding RNAs in human ovarian cancer cells with different metastatic potentials. Cancer Biol Med 10(3):138–141
Qiu JJ et al (2014) Expression and clinical significance of estrogen-regulated long non-coding RNAs in estrogen receptor alpha-positive ovarian cancer progression. Oncol Rep 31(4):1613–1622
Zeng S et al (2019) Upregulation of lncRNA AB073614 functions as a predictor of epithelial ovarian cancer prognosis and promotes tumor growth in vitro and in vivo. Cancer Biomark 24(4):421–428
Gao J et al (2015) Long non-coding RNA growth arrest-specific transcript 5 is involved in ovarian cancer cell apoptosis through the mitochondria-mediated apoptosis pathway. Oncol Rep 34(6):3212–3221
Li J et al (2017) LncRNA GAS5 suppresses ovarian cancer by inducing inflammasome formation. Biosci Rep. https://doi.org/10.1042/BSR20171150
Ma N et al (2018) Long non-coding RNA GAS5 inhibits ovarian cancer cell proliferation via the control of microRNA-21 and SPRY2 expression. Exp Ther Med 16(1):73–82
Feng J et al (2019) A noncoding RNA LINC00504 interacts with c-Myc to regulate tumor metabolism in colon cancer. J Cell Biochem 120(9):14725–14734
Yu Y et al (2017) A novel mechanism of lncRNA and miRNA interaction: CCAT2 regulates miR-145 expression by suppressing its maturation process in colon cancer cells. Mol Cancer 16(1):155
Xu JH et al (2018) The mRNA, miRNA and lncRNA networks in hepatocellular carcinoma: an integrative transcriptomic analysis from Gene Expression Omnibus. Mol Med Rep 17(5):6472–6482
Wei C et al (2017) Differentially expressed lncRNAs and miRNAs with associated ceRNA networks in aged mice with postoperative cognitive dysfunction. Oncotarget 8(34):55901–55914
Gao Y et al (2015) LncRNA-HOST2 regulates cell biological behaviors in epithelial ovarian cancer through a mechanism involving microRNA let-7b. Hum Mol Genet 24(3):841–852
Lei R et al (2017) Long noncoding RNA MALAT1-regulated microRNA 506 modulates ovarian cancer growth by targeting iASPP. Onco Targets Ther 10:35–46
Funding
None.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Ethical approval
The study protocol was approved by the Ethics Committee of Heze Municipal Hospital. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
All participants in this study were informed and gave a written consent.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, Y., He, X., Chen, Y. et al. Long non-coding RNA LINC00504 regulates the Warburg effect in ovarian cancer through inhibition of miR-1244. Mol Cell Biochem 464, 39–50 (2020). https://doi.org/10.1007/s11010-019-03647-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11010-019-03647-z